Conservación











Knowledge shortfalls and the effect of wildfireson biodiversity conservation in Guanajuato, Mexico
Tania Escalante a, *, Michelle Farfán b, Oscar Campos a, Leticia M. Ochoa-Ochoa c, Karen Flores-Quintal a, Diego R. García-Vélez a, Ana L. Medina-Bárcenas a, Fernando Saenz a
a Universidad Nacional Autónoma de México, Facultad de Ciencias, Grupo de Biogeografía de la Conservación, Circuito Exterior s/n, Ciudad Universitaria, Coyoacán, 04510 Mexico City, Mexico
b Universidad de Guanajuato, Campus Guanajuato, División de Ingenierías, Departamento de Ingeniería Geomática e Hidráulica, Av. Juárez No. 77, Zona Centro, 36000 Guanajuato, Guanajuato, Mexico
c Universidad Nacional Autónoma de México, Facultad de Ciencias, Departamento de Biología Evolutiva, Museo de Zoología “Alfonso L. Herrera”, Circuito Exterior s/n, Ciudad Universitaria, Coyoacán, 04510 Mexico City, Mexico
*Corresponding author: tescalante@ciencias.unam.mx (T. Escalante)
Received: 01 August 2023; accepted: 29 February 2024
Abstract
Knowledge of shortfalls could modify the geographic distribution patterns and limit the actions to conserve the biodiversity, even in the taxa best known. In addition, forest fires also could modify those patterns, but the potential effects of both factors have not been tested. Our aim was to analyze the effect of the Linnean and Wallacean shortfalls in the first evaluation of wildfire impacts on 22 amphibian and 13 mammal species distributed in Guanajuato, Mexico. We evaluated those shortfalls using the non-parametric estimator Chao2 and the Qs estimator and through maps of species richness patterns. To evaluate the effects of wildfires, we produced a fire recurrence map and quantified the burned area within species distributions and in 24 Protected Natural Areas (PNA) in the state. The Linnean shortfall showed some species missing to record in Guanajuato for both taxa, while the Wallacean shortfall showed poor quality of knowledge. Fire recurrence was high within 5 PNA. The richness patterns affected by fires covered nearly 17% of the surface of Guanajuato. Improving the knowledge of biogeographical patterns could provide better tools to stakeholders to decrease the negative impact of fires within PNA.
Keywords: Fire; Patterns; Priorities; Richness; Species distribution models
© 2024 Universidad Nacional Autónoma de México, Instituto de Biología. Este es un artículo Open Access bajo la licencia CC BY-NC-ND
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Déficits de conocimiento y el efecto de los incendios forestales en la conservación de la biodiversidad en Guanajuato, México
Resumen
Los déficits en el conocimiento podrían modificar los patrones de distribución geográfica y limitar las acciones para conservar la biodiversidad, incluso en taxones bien conocidos. Además, los incendios forestales también pueden modificar esos patrones, pero los efectos potenciales de ambos no han sido probados. Nuestro objetivo fue analizar el efecto de los déficits Linneano y Wallaceano en la primera evaluación de los impactos de los incendios forestales en 22 especies de anfibios y 13 de mamíferos en Guanajuato, México. Evaluamos esos déficits utilizando los estimadores Chao2 y Qs y con mapas de riqueza de especies. Para evaluar los efectos de incendios forestales, elaboramos un mapa de recurrencia de incendios y cuantificamos el área quemada dentro de las distribuciones de las especies y en 24 áreas naturales protegidas (ANP). El déficit Linneano mostró que faltan algunas especies por registrar para ambos taxones, mientras que el déficit Wallaceano mostró una mala calidad de conocimiento. La recurrencia de incendios fue alta dentro de 5 ANP. Los patrones de riqueza afectados por los incendios cubrieron cerca de 17% de la superficie de Guanajuato. Mejorar el conocimiento de los patrones biogeográficos brindará mejores herramientas para disminuir el impacto de los incendios dentro de las ANP.
Palabras clave: Fuego; Patrones; Prioridades; Riqueza; Modelos de distribución de especies
Introduction
Terrestrial vertebrates are among the best known taxonomic groups, and it is assumed that their distributional areas and their biogeographic patterns are equally well known. However, there are shortfalls that could mask the distributional patterns and therefore, bias the actions to conserve those patterns. Linnean and Wallacean shortfalls affect our knowledge and lead to inaccurate representations of the species richness patterns in taxonomic groups that are presumed to be well known. The Linnean shortfall refers to the discrepancy between formally described species and the number of species that actually exist, while the Wallacean shortfall is the lack of knowledge about the geographical distribution of the species (Hortal et al., 2015; Lomolino, 2004). Both the Linnean and the Wallacean shortfalls can be difficult to evaluate, and they are rarely quantified in the literature prior to a conservation prioritization analysis. For example, to our knowledge, they have never been taken into account when analyzing the effects of wildfires on biodiversity.
Throughout the history of the Earth, fire has been a natural process that has driven the configuration of ecosystems and the maintenance of biodiversity around the world (He et al., 2019; Kelly et al., 2020). Indeed, there are many terrestrial ecosystems that are prone to fire and whose composition and structure are controlled by fire, leading to their classification as fire-adapted ecosystems (He et al., 2019; Schlisky et al., 2007). However, the forest fire regime has been altered by human dynamics associated with fire management and land use change at local and global scales (Chuvieco et al., 2008; Farfán et al., 2018; Martínez-Torres et al., 2015). Several authors agree that there is an increase in the occurrence of wildfires globally (Kelly et al., 2020). Places that did not burn naturally are now burning; examples include the tropical forests of Southeast Asia (Chisholm et al., 2016) and South America (Barlow et al., 2020) to the tundra of the Arctic Circle (Hu et al., 2015). Given the magnitude at which fires are occurring, it has even been proposed that the current era should be coined the Pyrocene, the “age of fire” (Pyne, 2021). This has led to the current situation in which frequency and intensity of forest fires pose a threat to biodiversity conservation worldwide and to human societies; this is due to the damage they cause but also by contributing to global warming.
In Mexico, the effects of wildfires on the fauna are poorly documented. Salazar et al. (2019) proposed a map (scale 1:50,000) of the severity of the fires in the state of Guanajuato for 2017, 2018 and 2019, by calculating the area of burned forest within each of 3 degrees of damage severity: low moderate, high moderate and high. They estimated the total burned area in Guanajuato at 8,460 ha in 2017; 19,589 ha in 2018; and 52,713 ha in 2019 (Salazar et al., 2019). Recently, Farfán et al. (2021) produced a map predicting the occurrence of fires in Guanajuato based on climatic variables under ENSO conditions using a spatial model. They observed that wildfires do not occur in random locations; rather, they are more likely to occur when fragmented forest is immersed in an agricultural matrix, as is frequently the case in the southern part of the state (Farfán et al., 2021).
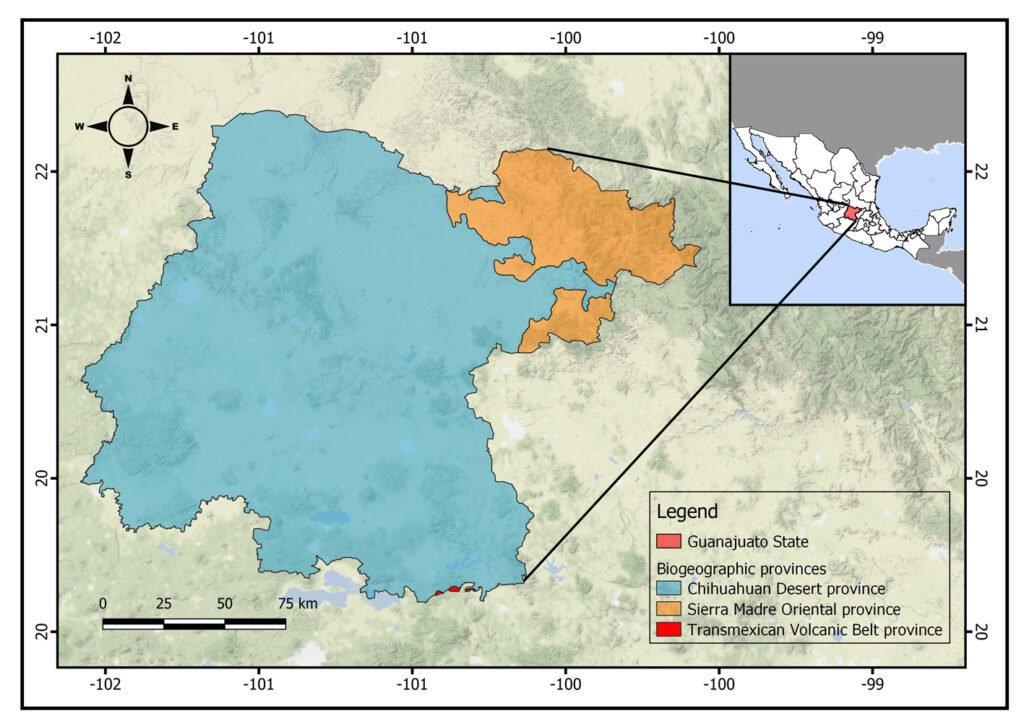
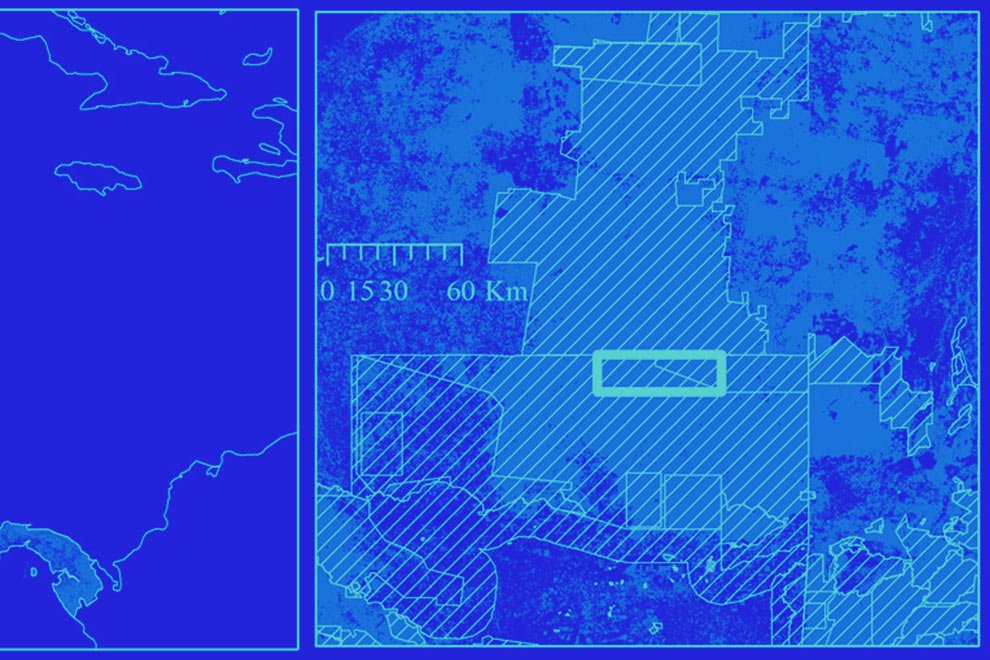
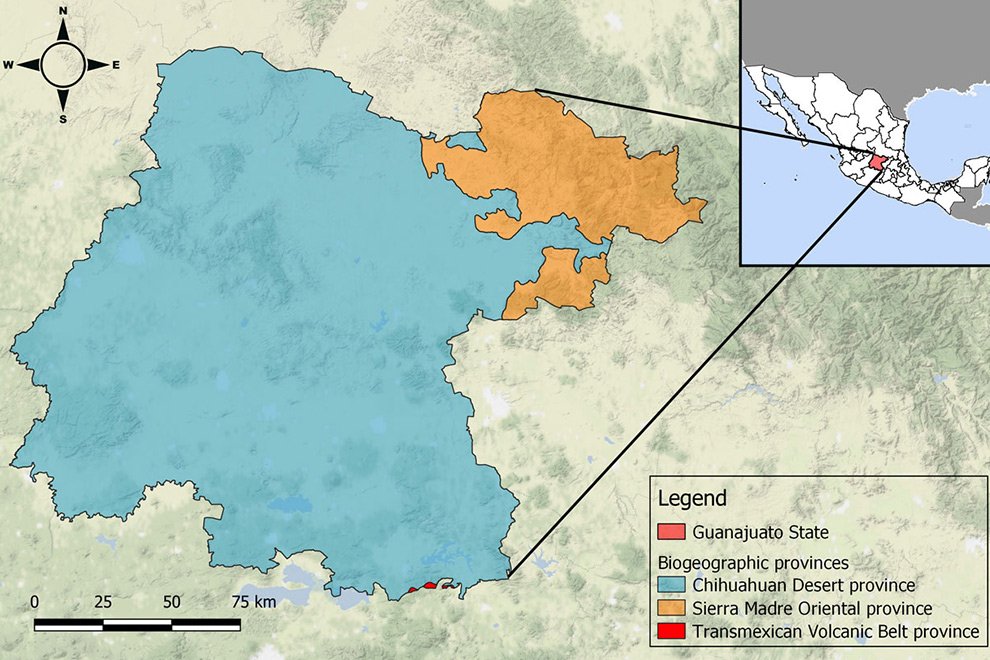
Figure 1. Location of the state of Guanajuato, Mexico, showing the biogeographic provinces.
Amphibians have been recognized as the most threatened terrestrial vertebrate class (Luedtke et al., 2023), and mammals could have significant declination in their populations due to fire, like in Australia (Geary et al., 2023). However, responses of animals to fire could be diverse because they are strongly related to their life-history traits (González et al., 2021). The responses of the amphibian species are variable and incompletely understood (Pilliod et al., 2003); while even among mammalian species, the effect of fire could be not consistent (González et al., 2021). Therefore, the effect of wildfires on distributional patterns of these taxa could be different.
In this study, our aim is to analyze the Linnean and Wallacean shortfalls in the context of the first evaluation of wildfire impacts on biodiversity. We use species of amphibians and mammals of Guanajuato, Mexico, as a study model and use the results to propose priority areas for conservation.
Material and methods
The state of Guanajuato is located in central Mexico, at 19°55’- 21°51’ N, 99°40’ – 102°06’ W. Most of the state is in the Chihuahuan Desert province, with a few areas in Sierra Madre Oriental and Transmexican Volcanic Belt provinces (Morrone et al., 2017; Fig. 1). Guajanuato has 24 Protected Natural Areas (PNA) mainly located in the southwest and center of the state. The largest of these areas is a biosphere reserve (Sierra Gorda), and the other PNA have a range of different levels of protection and activities allowed (SMAOT, 2022).
Although Guanajuato is not among the most biodiverse states in Mexico, it harbors a wide variety of ecosystems, from Pinus and Quercus forests to xerophytic scrubs (Conabio, 2012). Unfortunately, the extent of the agricultural and livestock areas, the high population density, the expansion of urban areas, and industrial activities have contributed to the destruction and disappearance of the original vegetation (Conabio, 2012).
Guanajuato harbors a total of 27 reported species of amphibians and 93 species of mammals, 8 and 25 of which, respectively, have been described as conservation priorities (DOF, 2010; Leyte-Manrique et al., 2022; Sánchez et al., 2016). Of all the species of amphibians and mammals inhabiting Guanajuato, we chose 22 species of amphibians and 13 species of mammals (Table 1) based on the following criteria: 1) valid nomenclature and at least one point record located in Guanajuato in the consulted databases (Flores-Villela & Ochoa-Ochoa, 2020; Escalante et al., 2018; GBIF.org, 2020a-ah); 2) geographic distribution mostly contained within Mexico; 3) at least 25 point records throughout the whole distribution in order to generate reliable species distribution models (SDM); and 4) considered conservation priorities.
In order to evaluate the Linnean and Wallacean shortfalls for those species, we searched the aforementioned databases for all valid point records in Guanajuato. These were initially overlapped to a grid of 0.25º latitude × 0.25º longitude, in QGIS v.3.16.16 (QGIS Development Team, 2020), which will be referred to hereafter as the “state scale”. We built a presence-absence matrix using the generated SDM.
To evaluate the Linnean shortfall based on point records, we quantified the observed richness (Sobs) as the recorded number of species of amphibians and mammals in each quadrat of 0.25º based on locality records. Then, we used the non-parametric estimator Chao2 in R (Kindt & Coe, 2005), to estimate the asymptotic richness of the incidence dataset (Gotelli & Colwell, 2011). The estimated richness Sest in Chao2 was obtained by the formula (Colwell & Coddington, 1994): Sest = Sobs + L2/(2M), where L = number of species that occur in only 1 quadrat, and M = number of species that occur in exactly 2 quadrats. Thus, this calculation provided a measure of how well the richness of each taxonomic group is known for those areas. We also performed the same analysis based on the species distribution models to explore how well the Linnean shortfall was corrected.
Table 1
List of species of amphibians and mammals in Guanajuato, Mexico, and data used in the analyses. Total records = number of point records after the nomenclatural and geographic validation (state scale). Records in Guanajuato = number of point records of each species into the geopolitical boundaries of the state of Guanajuato. Filtered records = subset of point records after the filter of 10 km applied to the total records. Records for modeling = subset of the filtered records used for model training. pROC = ROC partial of the best model.
| Species | Total records | Records in Guanajuato | Filtered records | Records for modeling | pROC |
| Amphibians | |||||
| Ambystoma velasci | 259 | 45 | 247 | 23 | 1.81 |
| Anaxyrus compactilis | 561 | 31 | 488 | 41 | 1.72 |
| Anaxyrus punctatus | 3,798 | 29 | 3,405 | 226 | 1.76 |
| Aquiloeurycea cephalica | 258 | 2 | 227 | 18 | 1.74 |
| Craugastor augusti | 599 | 16 | 508 | 54 | 1.58 |
| Dryophytes arenicolor | 2,514 | 138 | 2,269 | 161 | 1.81 |
| Dryophytes eximius | 820 | 19 | 761 | 72 | 1.75 |
| Eleutherodactylus guttilatus | 136 | 20 | 109 | 12 | 1.79 |
| Eleutherodactylus nitidus | 673 | 5 | 556 | 48 | 1.68 |
| Eleutherodactylus verrucipes | 237 | 2 | 167 | 13 | 1.52 |
| Hypopachus variolosus | 1,323 | 10 | 1,122 | 116 | 1.74 |
| Incilius nebulifer | 5,585 | 13 | 5,200 | 256 | 1.83 |
| Incilius occidentalis | 1,620 | 81 | 1,342 | 114 | 1.71 |
| Isthmura belli | 245 | 1 | 232 | 18 | 1.64 |
| Lithobates berlandieri | 3,747 | 51 | 3,401 | 247 | 1.56 |
| Lithobates megapoda | 127 | 9 | 101 | 12 | 1.65 |
| Lithobates montezumae | 696 | 94 | 598 | 51 | 1.75 |
| Lithobates neovolcanicus | 349 | 51 | 298 | 32 | 1.68 |
| Lithobates spectabilis | 544 | 3 | 416 | 40 | 1.64 |
| Rheohyla miotympanum | 383 | 1 | 329 | 27 | 1.60 |
| Smilisca baudinii | 4,381 | 2 | 3,590 | 277 | 1.68 |
| Spea multiplicata | 2,251 | 38 | 2,008 | 196 | 1.79 |
| Total | 31,090 | 661 | 27,374 | 2,054 | |
| Mammals | |||||
| Choeronycteris mexicana | 578 | 5 | 384 | 72 | 1.71 |
| Corynorhinus mexicanus | 204 | 4 | 149 | 28 | 1.79 |
| Dipodomys ornatus | 85 | 2 | 61 | 12 | 1.78 |
| Table 1. Continued | |||||
| Species | Total records | Records in Guanajuato | Filtered records | Records for modeling | pROC |
| Leptonycteris nivalis | 263 | 1 | 179 | 34 | 1.66 |
| Leptonycteris yerbabuenae | 575 | 9 | 370 | 70 | 1.75 |
| Lepus callotis | 199 | 6 | 150 | 28 | 1.75 |
| Peromyscus melanotis | 601 | 3 | 227 | 42 | 1.84 |
| Peromyscus difficilis | 897 | 10 | 453 | 85 | 1.69 |
| Peromyscus melanophrys | 594 | 25 | 372 | 70 | 1.59 |
| Rhogeessa alleni | 55 | 5 | 46 | 8 | 1.60 |
| Sciurus oculatus | 123 | 34 | 73 | 14 | 1.45 |
| Sigmodon leucotis | 103 | 9 | 72 | 14 | 1.50 |
| Sorex saussurei | 168 | 1 | 93 | 18 | 1.8 |
| Total | 4,445 | 114 | 2,629 | 495 |
To quantify the Wallacean shortfall, we used the Qs estimator (Murguía-Romero & Villaseñor, 2000), which is a measure of the quality of the records. QS can take values between ‘0’ and ‘1’ and is defined as (Murguía-Romero & Villaseñor, 2000): QS = F/[Sobs m/(1 – Es) – max (Sobs, m)], where F = the sum of frequencies of all classes multiplied by all classes (that is, the sum of all ‘1’ in the matrix); Es = measure of the proportion of the known richness related to the estimated richness; and m = the total number of quadrats. Murguía-Romero and Villaseñor (2000) characterized QS values above 80% as “very good”, values between 50% to 80% as “good” and less than 50% as “poor” data quality.
To compare the possible effect of the Wallacean shortfall in the biogeographic patterns, we performed a species distribution model (SDM) for each species in order to obtain a map of richness patterns for amphibians and mammals. Following the BAM diagram of Soberón and Peterson (2005), where the M corresponds to the region that is reachable by the species from established distributional areas in ecological time (Soberón & Peterson, 2005). The M for each species was obtained using the concept of extent of occurrence, defined as “the area contained within the shortest continuous imaginary boundary that can be drawn to encompass all the known, inferred or projected sites of present occurrence of a taxon, excluding cases of vagrancy” (IUCN, 2001). Therefore, we defined M as the area within a minimum convex hull polygon for each species constructed in QGIS v.3.16.16 (QGIS Development Team, 2020).
The M of each species was used to crop the 19 environmental layers of WorldClim 2 (Fick & Hijmans, 2017) and 3 topographic variables (slope, elevation and aspect; USGS, 2021) at ~ 1 km2 of resolution. The data points were filtered in Wallace software (Kass et al., 2018) to a distance of 10 km between points to reduce spatial biases, and retain useful information (Aiello-Lammens et al., 2015; Pearson et al., 2007). To avoid collinearity among the 22 variables for each species, we obtained the VIF (Mandeville, 2008; Montgomery & Peck, 1992), applying the packages usdm (Naimi et al., 2014) and rgdal (Bivand et al., 2015) in RStudio (RStudio Team, 2020).
The models were performed in the maximum entropy package kuenm (Cobos et al., 2019) in RStudio (RStudio Team, 2020). The occurrence dataset for each species was divided as follows: 75% of the points were used for training and 25% for testing; and a set of independent occurrences of 25% for a last evaluation; those datasets were built with the kuenm_occsplit function in kuenm. For the next step, we used the function kuenm_cal, using the feature classes: linear, quadratic and hinge; and the regularization multipliers 0.5, 1, 1.5, 2, 2.5, 3, 3.5 and 4. All models were evaluated with kuenm_ceval, calculating the ROC partial with E = 10 (Peterson et al., 2008), and Akaike criterion for small samples (AICc; Warren & Seifert, 2011). The final best model for each species was obtained on a clog-log scale using the pROC value in NicheToolbox (Osorio-Olvera et al., 2020).
To produce binary maps of geographic distribution area of each species, the final best model was reclassified using the “10th percentile training presence” threshold, and cropped to the political boundaries of Guanajuato.
Wildfires. We used hotspot data from the MODIS sensor at a resolution of 1 km2 for the years 2000 to 2021, downloaded from NASA Earth Data Cloud (2020). Each hotspot was overlapped to a net of ~ 1 km2 covering the state of Guanajuato in QGIS (QGIS Development Team, 2020), and we counted the number of hotspots in each square of the net. This area will be referred to as the “fine scale”.
In order to compare the number of fires in each square of the net with the scale of the models, we transformed this number between ‘0’ and ‘1’ through a min-max normalization (Farrús et al., 2007; Jain et al., 2005). This procedure was useful to evaluate the effect of the wildfires on the SDM of each species. All cartographic products were projected to UTM zone 14 north coordinates, which corresponds to the state of Guanajuato. To quantify the impact of the fires on species richness patterns, we rasterized the map of the number of fires, from which we produced a new map of kernel density with a radius of 3,000 m, using the software DINAMICA EGO (Ferreira et al., 2019). The map of kernel density was multiplied by the richness map for each taxonomic group. Finally, we quantified the burned area for the maps and for the Protected Natural Areas (SMAOT, 2022).
Results
Shortfalls. We obtained 31,090 records of amphibians considering the whole distribution for the 22 amphibian species, and 661 records within Guanajuato. For the 13 species of mammals, the number of records was 74 in Guanajuato and 2,629 in the whole distribution. The species with the most records was the coastal plain toad Incilius nebulifer (5,585 records) for the amphibians and the southern rock mouse Peromyscus difficilis (453 records; see Table 1) for the mammals. However, within Guanajuato, there were only 13 and 9 records for these species, respectively. The species with the highest number of point records within Guanajuato were the canyon tree frog Dryophytes arenicolor with 138 records, and the Peters’s squirrel Sciurus oculatus, with 34 records.
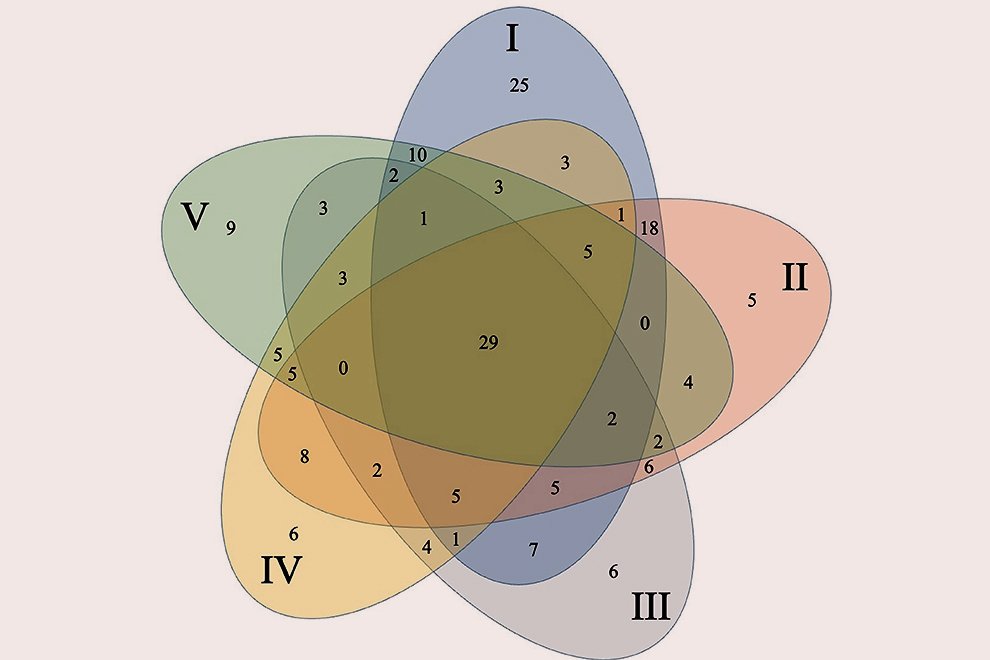
The richest 0.25º quadrat for amphibians had 11 species and the quadrat with the most mammals had 5 species, while the lowest number of species per quadrat was 1 for both taxa, although there were a few marginal quadrats without data for amphibians and some complete quadrats without data for mammals (~ 12). The maps of quadrats with the observed richness (Sobs in Chao2) for both groups are shown in the Supplementary material: figures 1S, 2S. The main results of the Chao2 estimator are shown in Table 2. For both groups, the number of observed species (Sobs) was lower than the expected number (Sest) for Guanajuato. In the case of the Chao2 estimated with species distribution models, the species richness was the same as the expected species.
Regarding the Wallacean shortfall, the Qs estimator had a value of 23% for the amphibians and 28% for mammals at the state scale. These values were categorized as “poor” quality data in both taxa, but Qs was worse in amphibians.
Table 2
Results of the recorded and estimated richness for the complete distributional data of amphibians and mammals of Guanajuato, Mexico based both on species richness and on species distribution models (SDM).
| Taxonomic group | Recorded richness (Sobs) | Estimated richness (Sest) | Standard deviation (SD) | Estimated richness with SDM (Sest) |
| Amphibians | 22 | 24.61 | 3.42 | 24 |
| Mammals | 13 | 14.94 | 3.64 | 14 |

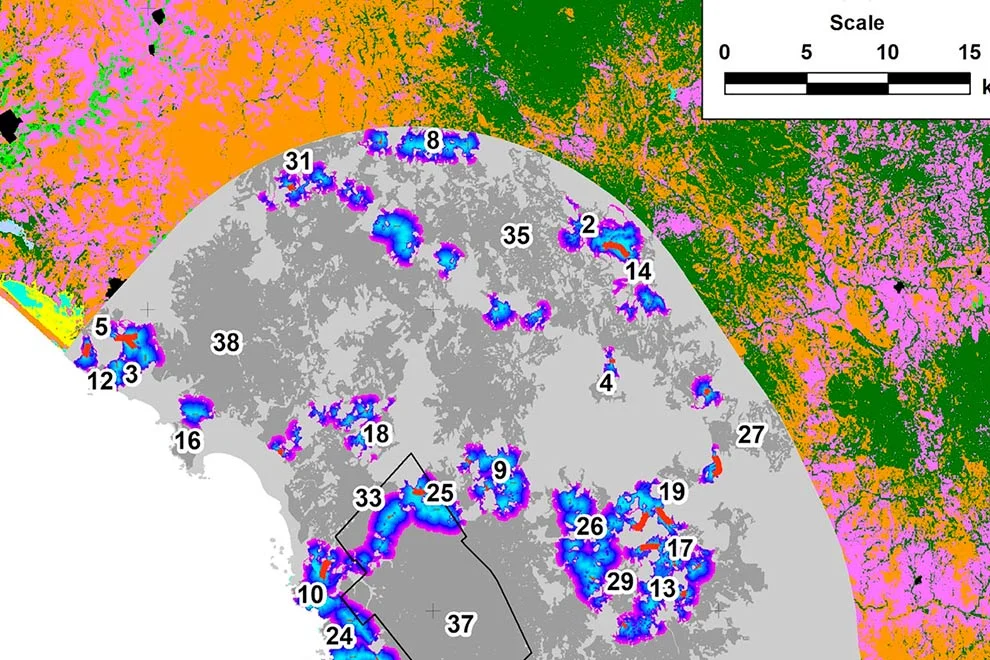
Figure 2. Richness pattern for 22 modeled species of amphibians and the Protected Natural Areas in Guanajuato, Mexico (black polygons): 1. Palenque, 2. Peña Alta, 3. Sierra de Pénjamo, 4. Sierra de los Agustinos, 5. Las Fuentes, 6. Sierra de Lobos, 7. Las Musas, 8. Lago Cráter La Joya, 9. Cerro de los Amoles, 10. Parque Metropolitano, 11. Cerro de Arandas, 12. Cuenca Alta del Río Temascatio, 13. Cerro del Cubilete, 14. Cerros El Culiacán y La Gavia, 15. Cuenca de La Esperanza, 16. Mega Parque de la Ciudad de Dolores Hidalgo, 17. Presa La Purísima y su Zona de Influencia, 18. Presa de Neutla y su Zona de Influencia, 19. Laguna de Yuriria y su Zona de Influencia, 20. Pinal de Zamorano, 21. Región Volcánica Siete Luminarias, 22. Sierra Gorda de Guanajuato, 23. Cuenca de la Soledad, 24. Presa de Silva y Áreas Aledañas.
Some results of the modeling process are shown in Table 1. The distribution models of amphibians predicted that the species Craugastor augusti, Dryophytes arenicolor, Dryophytes eximius, Incilius occidentalis, Lithobates montezumae, Lithobates neovolcanicus, and Spea multiplicata are distributed in more than 90% of the surface of Guanajuato. Meanwhile, the species Aquiloeurycea cephalica (31%), Smilisca baudini (25%), Lithobates berlandieri (19%), and Incilius nebulifer (3%) had the lowest proportion of distribution in Guanajuato. Regarding the mammals, Choeronycteris mexicana, Corynorhynus mexicanus, Leptonycteris nivalis, Lepus callotis, Peromyscus difficilis, Peromyscus melanophrys, and Peromyscus melanotis were the most widely distributed in Guanajuato (more than the 90% of the state is predicted as part of their distribution), while Sorex saussurei was the only species with a proportion less than 30%.
For the richness patterns, the pixels with highest number of species modeled to be present were similar between amphibians and mammals at the fine scale (Figs. 2, 3), showing a diagonal strip of high richness from the northwest to the southeast, which coincided with 14 PNA: Las Fuentes, Sierra de Lobos, Parque Metropolitano, Cuenca Alta del Río Temascatío, Cerro del Cubilete, Cuenca de La Esperanza, Presa de Neutla, Cuenca de la Soledad, Sierra de los Agustinos, Lago Cráter La Joya, Cerro de los Amoles, Cerros El Culiacán y La Gavia, Laguna de Yuriria and Región Volcánica Siete Luminarias. There were also other sites of high diversity, for example within the PNA of Sierra de Pénjamo and Sierra Gorda.
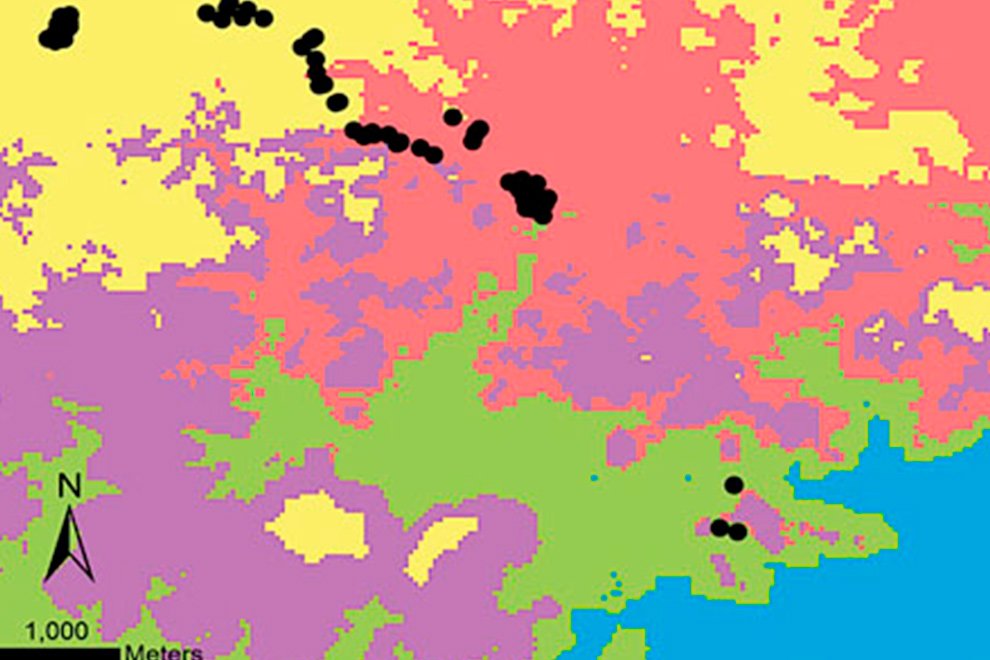
Wildfires. The maximum number of wildfires per quadrant at the fine scale was 6. To further explore the risk of wildfire recurrence, we built a risk map using the recurrence of fires in each square, with 3 classes: 1) low risk, for pixels with one fire during the analyzed period; 2) medium risk, for pixels with 2 or 3 fires; and 3) high risk, for pixels with 4, 5 or 6 fires (Fig. 4). A large proportion of quadrats with high recurrence of fires occurred outside PNA (for example at northern Guanajuato), but there were also some high risk zones within PNA, like Palenque, Peña Alta, Sierra de Pénjamo, Las Musas and Región Volcánica Siete Luminarias.

Figure 3. Richness pattern for 13 modeled species of mammals and the Protected Natural Areas in Guanajuato, Mexico (black polygons): 1. Palenque, 2. Peña Alta, 3. Sierra de Pénjamo, 4. Sierra de los Agustinos, 5. Las Fuentes, 6. Sierra de Lobos, 7. Las Musas, 8. Lago Cráter La Joya, 9. Cerro de los Amoles, 10. Parque Metropolitano, 11. Cerro de Arandas, 12. Cuenca Alta del Río Temascatio, 13. Cerro del Cubilete, 14. Cerros El Culiacán y La Gavia, 15. Cuenca de La Esperanza, 16. Mega Parque de la Ciudad de Dolores Hidalgo, 17. Presa La Purísima y su Zona de Influencia, 18. Presa de Neutla y su Zona de Influencia, 19. Laguna de Yuriria y su Zona de Influencia, 20. Pinal de Zamorano, 21. Región Volcánica Siete Luminarias, 22. Sierra Gorda de Guanajuato, 23. Cuenca de la Soledad, 24. Presa de Silva y Áreas Aledañas.
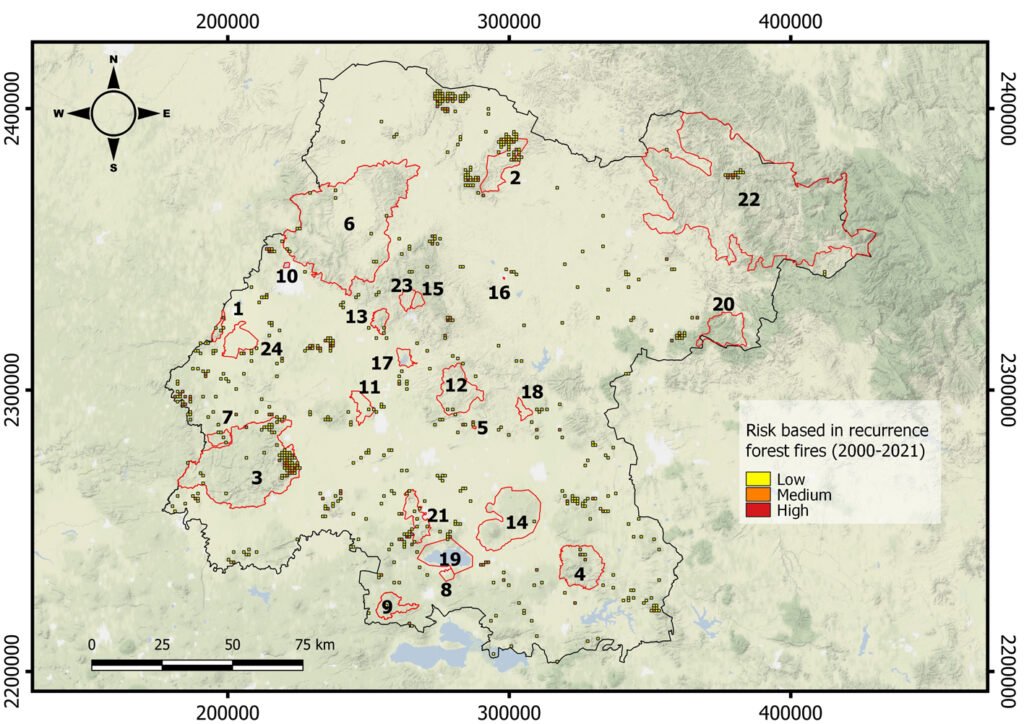
Figure 4. Map of fire risk in Guanajuato between the years 2000 and 2021, based on the recurrence of fires in a 1 km square, with 3 classes: (1) low risk, for squares with one fire during the analyzed period; (2) medium risk, for squares with 2 or 3 fires; and (3) high risk, for squares with 4, 5 or 6 fires. Red polygons represent the Protected Natural Areas.
Table 3
Potential distribution area predicted by the modeling for 22 amphibian and 13 mammal species and quantification of habitat lost due to wildfires relative to the total area of Guanajuato, Mexico (30,702 km2).
| Species | Surface of potential distribution area occupied in Guanajuato (km2) | Percentage of potential distribution area occupied (%) | Surface of potential distribution area affected by wildfires (km2) | Percentage of potential distribution area affected by wildfires (%) |
| Amphibians | ||||
| Ambystoma velasci | 22,351 | 73 | 3,620 | 16 |
| Anaxyrus compactilis | 26,061 | 85 | 4,948 | 19 |
| Anaxyrus punctatus | 18,241 | 59 | 2,440 | 13.3 |
| Aquiloeurycea cephalica | 9,653 | 31 | 1,246 | 12.9 |
| Craugastor augusti | 30,702 | 100 | 5,264 | 17.1 |
| Dryophytes arenicolor | 30,696 | 99.9 | 5,262 | 17.1 |
| Dryophytes eximius | 30,267 | 98.5 | 5,249 | 17.3 |
| Eleutherodactylus guttilatus | 21,636 | 70 | 2,088 | 14.2 |
| Eleutherodactylus nitidus | 22,888 | 75 | 4,663 | 20.3 |
| Eleutherodactylus verrucipes | 20,519 | 67 | 2,756 | 13.4 |
| Hypopachus variolosus | 19,076 | 62 | 3,951 | 20.7 |
| Incilius nebulifer | 873 | 3 | 55 | 6.3 |
| Incilius occidentalis | 30,546 | 99 | 5,257 | 17.2 |
| Isthmura bellii | 26,647 | 87 | 4,925 | 18.4 |
| Lithobates berlandieri | 5,772 | 19 | 614 | 10.6 |
| Lithobates megapoda | 20,866 | 68 | 4,422 | 21.1 |
| Lithobates montezumae | 29,957 | 97.5 | 5,242 | 17.4 |
| Lithobates neovolcanicus | 28,670 | 93 | 5,137 | 17.9 |
| Lithobates spectabilis | 24,821 | 81 | 3,893 | 15.6 |
| Rheohyla miotympanum | 18,056 | 59 | 3,516 | 19.4 |
| Smilisca baudinii | 7,631 | 25 | 1,579 | 20.6 |
| Spea multiplicata | 30,661 | 99.8 | 5,264 | 17.1 |
| Mammals | ||||
| Choeronycteris mexicana | 30,608 | 99.69 | 5,242 | 17.12 |
| Corynorhynus mexicanus | 30,608 | 99.69 | 5,242 | 17.12 |
| Dipodomys ornatus | 18,553 | 60.43 | 3,429 | 18.48 |
| Leptonycteris nivalis | 30,608 | 99.69 | 5,242 | 17.12 |
| Leptonycteris yerbabuenae | 24,955 | 81.28 | 4,474 | 17.93 |
| Lepus callotis | 28,469 | 92.73 | 5,025 | 17.65 |
| Peromyscus difficilis | 30,142 | 98.18 | 5,225 | 17.34 |
| Peromyscus melanophrys | 29,606 | 96.43 | 5,227 | 17.65 |
| Peromyscus melanotis | 29,231 | 95.21 | 5,183 | 17.73 |
| Rhogessa alleni | 17,604 | 57.34 | 3,353 | 19.05 |
| Sciurus oculatus | 14,564 | 47.44 | 1,923 | 13.20 |
| Sigmodon leucotis | 18,215 | 59.33 | 3,139 | 17.23 |
| Sorex saussurei | 4,151 | 13.52 | 756 | 18.22 |

Figure 5. Richness map of amphibians intersected with the kernel density of recurrence of the wildfires in Guanajuato, Mexico. Black polygons represent the Protected Natural Areas.
1. Palenque, 2. Peña Alta, 3. Sierra de Pénjamo, 4. Sierra de los Agustinos, 5. Las Fuentes, 6. Sierra de Lobos, 7. Las Musas, 8. Lago Cráter La Joya, 9. Cerro de los Amoles, 10. Parque Metropolitano, 11. Cerro de Arandas, 12. Cuenca Alta del Río Temascatio, 13. Cerro del Cubilete, 14. Cerros El Culiacán y La Gavia, 15. Cuenca de La Esperanza, 16. Mega Parque de la Ciudad de Dolores Hidalgo, 17. Presa La Purísima y su Zona de Influencia, 18. Presa de Neutla y su Zona de Influencia, 19. Laguna de Yuriria y su Zona de Influencia, 20. Pinal de Zamorano, 21. Región Volcánica Siete Luminarias, 22. Sierra Gorda de Guanajuato, 23. Cuenca de la Soledad, 24. Presa de Silva y Áreas Aledañas.
Respect to the temporal distribution of the recurrence of forest fires, for the period of time analyzed, the years 2017, 2019, and 2021 had the highest number of fires (72, 110 and 150, respectively). The map of kernel density is shown in Supplementary material: Figure 3S. The map was transformed to a binary map and overlapped with the patterns of richness of amphibians and mammals, to produce the maps in figures 5, 6.
The evaluation of the effects of the wildfires on the model of each species is shown in Table 3. The mean percentage of burned potential distribution area was 16.49 and 17.37 for amphibians and mammals, respectively. Some species’ distribution areas were more strongly affected by fires, such as Eleutherodactylus nitidus, Hypopachus variolosus, Lithobates megapode, and Smilisca baudinii, all of which were amphibians for which more than 20% of their distribution area had been burned. For mammals, the most affected species was Rhogessa alleni (19.05%), followed by Sorex saussurei (18.22%).
The area of Guanajuato affected by fires measured nearly 5,200 km2 (17%; Figs. 5, 6), with high diversity zones for amphibians located in the northwest, near Sierra de Lobos (PNA 6) and Peña Alta (PNA 2). Other important affected areas for amphibians coincided with high recurrence of wildfires in the southern of Guanajuato in Región Volcánica Siete Luminarias (PNA 21), Sierra de los Agustinos (PNA 4) and Cerro de los Amoles (PNA 9). For mammals, some of the most strongly affected areas coincided with those of the amphibians (e.g., within Cerro de los Amoles; PNA 9), but there were also areas that were unique to mammals (Fig. 6). For example, there were areas of high mammal richness with wildfires in central Guanajuato, which did not coincide with any PNA, as well as southeastern areas in Sierra de los Agustinos (PNA 4). Fortunately, areas with high richness for both amphibians and mammals were not affected by fires, like Sierra de Lobos (PNA 6), Cuenca de la Esperanza (PNA 15), and Cuenca de la Soledad (PNA 23).
Discussion
Linnean shortfalls in the state of Guanajuato could have medium effects because the estimator predicted at least 2 additional species for each taxon relative to total currently known. This suggests that the current species inventories are not yet complete. This finding does not dismiss possible shortfalls at more detailed scales, because the number of records within the state of Guanajuato is very low, with an average of 30 per each species of amphibian and only 6 of each species of mammal. It would therefore be informative to carry out more specific analyses within the quadrats where 0 or 1 species were recorded. Increased collection effort in the field could improve the problems of undersampling, since the number of total data points in Guanajuato is very low for some species (v. gr. Aquiloeurycea cephalica with 2 records, and Leptonycteris nivalis with 1 record; Table 1). It is interesting to highlight that when performing the analyses with SDM, the Linnean shortfall is apparently corrected (Table 2). However, these results should be taken with caution, because it is possible that there are commission errors in models or that those areas actually correspond to sister species (Rodrigues et al., 2019; Acevedo et al., 2014).

Figure 6. Richness map of mammals intersected with the kernel density of recurrence of the wildfires in Guanajuato, Mexico. Black polygons represent the Protected Natural Areas.
1. Palenque, 2. Peña Alta, 3. Sierra de Pénjamo, 4. Sierra de los Agustinos, 5. Las Fuentes , 6. Sierra de Lobos, 7. Las Musas, 8. Lago Cráter La Joya, 9. Cerro de los Amoles, 10. Parque Metropolitano, 11. Cerro de Arandas, 12. Cuenca Alta del Río Temascatio, 13. Cerro del Cubilete, 14. Cerros El Culiacán y La Gavia, 15. Cuenca de La Esperanza, 16. Mega Parque de la Ciudad de Dolores Hidalgo, 17. Presa La Purísima y su Zona de Influencia, 18. Presa de Neutla y su Zona de Influencia, 19. Laguna de Yuriria y su Zona de Influencia, 20. Pinal de Zamorano, 21. Región Volcánica Siete Luminarias, 22. Sierra Gorda de Guanajuato, 23. Cuenca de la Soledad, 24. Presa de Silva y Áreas Aledañas.
On the other hand, the Wallacean shortfall was highly relevant for both amphibians and mammals, showing poor quality. Murguía-Romero and Villaseñor (2000) suggested that the quality of the records is related to the geographical resolution of the biogeographical analysis. In future analysis within the state of Guanajuato and using smaller quadrats (for example, close in size to the pixels of our models), the Wallacean shortfall could strongly affect the observed data, reaching very poor data quality. Thus, it seems to be the more important shortfall for these vertebrate species.
For our 25 species, probably the effect of the Linnean shortfall has a less dramatic effect than the Wallacean shortfall. There are multiple potential explanations for these shortfalls, including low intensity and spatial variation of sampling, which can directly affect biodiversity estimators like species richness (Oliveira et al., 2016). Continuing the study of these shortfall will be important for the correct implementation of conservation strategies, for example with other methods including correlations using the sampling effort (Oliveira et al., 2016), rate of descriptions and number of taxonomists (Joppa et al., 2011), many different algorithms of species distribution modeling and maps of ignorance (Oliveira et al., 2016; Rocchini et al., 2011; Tessarolo et al., 2021), among others.
In spite of the extant Wallacean shortfalls, the richness patterns for both taxa were partially recognized; specially, the recorded most richness quadrats at 0.25º also showed the modeled pattern for amphibians. For mammals, the highest richness area near to Cuenca de la Esperanza was identified also for a quadrat of 0.25º, coinciding with the models. Amphibians and mammals shared many (though not all) areas of high richness, generally following a northwest-southeast diagonal across the state. The partial similarity in those richness patterns could be useful in Systematic Conservation Planning, because both taxa could represent each other as good surrogates (Escalante et al., 2020).
In general, amphibians and mammals are over-represented taxa in databases, although amphibians are less represented than mammals (Troudet et al., 2017). Few articles have quantified the Linnean and Wallacean shortfalls prior to biogeographic analysis. In particular, Oliveira et al.(2016) suggested that terrestrial vertebrates have similar biases compared with some taxa of arthropods, contradicting the statement that terrestrial vertebrates are better suited for biogeographic and conservation studies. In some places, such as the state of Guanajuato, amphibians and mammals could have similar Linnean shortfalls, but differ in the severity of their Wallacean shortfalls, which could modify the biogeographic patterns identified.
The relevance of including shortfall analysis in biogeographical studies, mainly in those related to species conservation, lies in the fact that the Linnean and Wallacean shortfalls strongly influence the possible results, since the data on the identity and distribution of the species are crucial to identify patterns in biodiversity, as well as the processes that modify those patterns (Hortal et al., 2015). In particular, Wallacean shortfalls can also alter estimates of threatened conservation status, since range size is regularly used in conservation (Hortal et al., 2015). Species with small ranges have higher priority in many international and national standards (DOF, 2010; IUCN, 2012). Therefore, Wallacean shortfalls could lead to some taxa and areas being disproportionately prioritized over others because their distribution areas have been erroneously underestimated (Riddle et al., 2011). In addition, other shortfalls that potentially can affect the biogeographic patterns should be investigated, like Darwinian shortfall (Diniz-Filho et al., 2013, 2023), and even distinct categories of Linnean shortfalls (Vergara-Asenjo et al., 2023).
As we expected, wildfires affected all species, but in different ways. For amphibians, the species Smilisca baudinii, in addition to having a small distribution area in Guanajuato compared to the rest of the species, is one of the most affected by wildfires occurrences. These observations may suggest that Smilisca baudinii should be considered a priority species for conservation in the state of Guanajuato. Furthermore, we also highlight Lithobates megapoda, which is listed in the 2019 update of the NOM-059-SEMARNAT-2010 (DOF, 2019) as a species under special protection and described as sensitive to habitat degradation (Santos-Barrera & Flores-Villela, 2004).
On the other hand, in the northeastern part of the state, the effects seem to be minimal compared to the south and southwest, but it is important to remember that species with specific habitat requirements such as Incilius nebulifer, Lithobates berlandieri, and Smilisca baudinii are distributed in this area. Therefore, attention should be paid to investigating the sources of ignition present at this area in order to prevent future wildfires, since the loss of habitat could result in the disappearance of these species. Finally, in accordance with Clivillé et al.(1997), who describe the effect of fires on amphibians from 3 points of view (habitat, species and individual), our study only focuses on the effect of these events on the habitat. Thus, the effects on the distribution of the selected species can be interpreted as loss of habitat and vegetation cover, trophic resources and humidity due to wildfires, which are determining characteristics for the presence of amphibians and their reproduction (Clivillé et al., 1997).
For the case of mammals, Sorex saussurei (the Saussure’s shrew) is the species with the narrowest geographic distribution in Guanajuato. This shrew is only distributed in Mexico and Guatemala, and even though it is considered as least concern on the Red List of Threatened Species (IUCN, 2017), some populations in Mexico have been categorized as threatened and under special protection (DOF, 2019). Secondly, Peters’s squirrel Sciurus oculatus occupies less than 50% of the surface of Guanajuato and is also categorized as least concern in the Red List of Threatened Species (IUCN, 2016). Sciurus oculatus is found only within Mexico, and is under special protection in national legislation NOM-059-SEMARNAT-2010, update of 2019 (DOF, 2019). Both species face continuing decline in the extent and quality of habitat due to land use change (Conafor, 2020; IUCN, 2016, 2017), which is exacerbated by repeated burning episodes that decrease the area occupied by each species by 13-18%. According to Zamudio (2012), most of the plant communities in the state of Guanajuato have significantly changed in their structure, floristic composition and physiognomy. Consequently, their distribution areas have been gradually reduced. Currently, 63% of the territory has been transformed into agricultural areas, human settlements, and areas devoid of vegetation (Roth et al., 2016).
The recurrence of wildfires, mainly in the southern part of Guanajuato, represents an important threat to biodiversity conservation within PNA, which are surrounded by a complex matrix of rainfed and irrigated agricultural land uses. This result was also found by Farfán et al. (2020, 2021), where both the probabilities of anthropogenic ignition and climate under the ENSO climate conditions lead to high wildfire risk in this region of the state. These PNA urgently need fire management plans that can integrate fire prevention actions at the local level in the context of global warming. On a global scale, wildfires have been responsible for up to 27% of the loss of tree cover between 2001 and 2021 (Tyukavina et al., 2022). In Mexico alone, in 2021, 408.75 km2 of forest were lost due to fire (Tyukavina et al., 2022). The effects of wildfires on biodiversity patterns could be understimated if these shortfalls are underestimating the biodiversity. Therefore, actions at the international level are also urgent in order to prevent damage to unknown biogeographic patterns.
This is the first study for the state of Guanajuato and for Mexico that addresses the effect of wildfire on the potential distribution of 2 important taxonomic groups: amphibians and mammals. The evaluation of the Linnean and Wallacean shortfalls for any taxonomic group is essential before the identification of geographic patterns involved as criteria for conservation planning, even in terrestrial vertebrates which are assumed to be adequately sampled. The Wallacean shortfall could lead to underestimations of the effects of perturbations such as wildfires. This is particularly true of species that are already vulnerable due to anthropogenic factors such as land cover change, illegal trafficking, etc., as well as intrinsic factors like the size of their natural distributional areas, because it is unknown whether undersampling could represent geographically rare species. Improving the biogeographical knowledge of the patterns of amphibians and mammals could provide better tools to stakeholders in order to generate fire management plans to prevent the negative impact of the wildfire within protected areas around the world.
Acknowledgements
We thank Julián A. Velasco and Luis J. Aguirre for their help with the parametrization of Maxent, and Miguel Murgía for assistance with the quantification of the Qs estimator. We also thank to the anonymous reviewers and the Associate Editor for their careful reading of this manuscript.
References
Acevedo, P., Melo-Ferreira, J., Real, R., & Alves, P. C. (2014). Evidence for niche similarities in the allopatric sister species Lepus castroviejoi and Lepus corsicanus. Journal
of Biogeography, 41, 977–986. https://doi.org/10.1111/jbi.12
270
Aiello-Lammens, M. E., Boria, R. A., Radosavljevic, A., Vilela, B., & Anderson, R. P. (2015). spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography, 38,541–545. https://doi.org/10.1111/ecog.01132
Barlow, J., Berenguer, E., Carmenta, R. & França, F. (2020). Clarifying Amazonia’s burning crisis. Global Change Biology, 26, 319–321. https://doi.org/10.1111/gcb.14872op
Bivand, R., Keitt, T., Rowlingson, B., Pebesma, E., Sumner, M., Hijmans, R. et al. (2015). Package ‘rgdal’. Bindings for the Geospatial Data Abstraction Library. Retrieved on December, 2020 from: http://rgdal.r-forge.r-project.org
Chisholm, R. A., Wijedasa, L. S., & Swinfield, T. (2016). The need for long-term remedies for Indonesia’s forest fires. Conservation Biology, 30, 5–6. https://doi.org/10.1111/cobi.
12662
Chuvieco, E., Giglio, L. & Justice, C. (2008). Global characterization of fire activity: toward defining fire regimes from Earth observation data. Global Change Biology, 14, 1488–1502. https://doi.org/10.1111/j.1365-2486.2008.01585.x
Clivillé, S., Montori, A., Llorente, G. A., Santos, X., & Carretero, M. A. (1997). El impacto de los incendios forestales sobre los anfibios. Quercus, 138,10–13.
Cobos, M. E., Peterson, A. T., Barve, N., & Osorio-Olvera, L. (2019). Kuenm: An R package for detailed development of ecological niche models using Maxent. PeerJ, 7,e6281. https://doi.org/10.7717/peerj.6281
Colwell, R. K., & Coddington, J. A. (1994). Estimating terrestrial biodiversity through extrapolation. Philosophical Transactions of the Royal Society B, 345, 101–118. https://doi.org/10.1098/rstb.1994.0091
Conabio (Comisión Nacional para el Conocimiento y Uso de la Biodiversidad). (2012). La biodiversidad en Guanajuato: estudio de estado (Vol. II). Guanajuato, Mexico: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (Conabio)/ Instituto de Ecología del Estado de Guanajuato (IEE).
Conafor (Comisión Nacional Forestal). (2020). Estimación de la tasa de deforestación bruta en México para el periodo 2001-2018 mediante el método muestreo. Documento técnico. Comisión Nacional Forestal. Jalisco, México. Retrieved in 2022 from: http://www.conafor.gob.mx:8080/documentos/docs/1/7768Documento%20tecnico%202020%20Deforestacion%20Bruta%20Final.pdf
DOF (Diario Oficial de la Federación). (2010). NORMA
Oficial Mexicana NOM-059-SEMARNAT-2010, Protección ambiental-Especies nativas de México de flora y fauna silvestres-Categorías de riesgo y especificaciones para
su inclusión, exclusión o cambio-Lista de especies en
riesgo. Mexico City, Mexico. Retrieved on March 3rd, 2021 from: https://dof.gob.mx/nota_detalle_popup.php?
codigo=5173091
DOF (Diario Oficial de la Federación). (2019). Modificación del Anexo Normativo III, Lista de especies en riesgo de la Norma Oficial Mexicana NOM-059-SEMARNAT-2010, Protección ambiental-Especies nativas de México de flora y fauna silvestres-Categorías de riesgo y especificaciones para su inclusión, exclusión o cambio-Lista de especies en riesgo, publicada el 30 de diciembre de 2010. Secretaría de Gobernación (SEGOB). Mexico City, Mexico. Retrieved on March 03rd, 2021 from: https://www.dof.gob.mx/nota_detalle.php?codigo=5578808&fecha=14/11/2019#gsc.tab=0
Diniz-Filho, J. F, Jardim, L., Guedes, J. J., Meyer, L., Stropp, J., Frateles, L. E. et al. (2023). Macroecological links between the Linnean, Wallacean, and Darwinian shortfalls. Frontiers of Biogeography, 15, e59566. http://dx.doi.org/10.21425/F5FBG59566
Diniz-Filho, J. A. F., Loyola, R. D., Raia, P., Mooers, A. O., & Bini, L. M. (2013) Darwinian shortfalls in biodiversity conservation. Trends in Ecology and Evolution, 28, 689–695. https://doi.org/10.1016/j.tree.2013.09.003
Escalante, T., Noguera-Urbano, E. A., & Corona, W. (2018). Track analysis of the Nearctic region: Identifying complex areas with mammals. Journal of Zoological Systematics and Evolutionary Research, 56, 466–477. https://doi.org/10.1111/jzs.12211
Escalante, T., Varela-Anaya, A. M., Noguera-Urbano, E. A., Elguea-Manrique, L. M., Ochoa-Ochoa, L. M., Gutiérrez-Velázquez, A. L. et al. (2020). Evaluation of five taxa as surrogates for conservation prioritization in the Trans-
mexican Volcanic Belt, México. Journal for Nature Conservation, 54, e125800. https://doi.org/10.1016/j.jnc.20
20.125800
Farfán, M., Pérez-Salicrup, D. R., Flamenco-Sandoval, A., Nicasio-Arzeta, S., Mas, J. F., & Ramírez-Ramírez, I. (2018). Modeling anthropic factors as drivers of wildfire occurrence at the Monarch Butterfly Biosphere. Madera y Bosques, 24, 1–15. https://doi.org/10.21829/myb.2018.2431591
Farfán, M., Flamenco-Sandoval, A., Rodríguez-Padilla, C. R., Rodrigues-de Sousa Santos, L., González-Gutiérrez, I., & Gao, Y. (2020). Cartografía de la probabilidad de ocurrencia a incendios forestales para el estado de Guanajuato: Una aproximación antrópica de sus fuentes de ignición. Acta Universitaria, 30, 1–15. https://doi.org/10.15174/au.2020.2953
Farfán, M., Domínguez, C., Espinoza, A., Jaramillo, A., Alcántara, C., Maldonado, V. et al. (2021). Forest fire probability under ENSO conditions in a semi-arid region: a case study in Guanajuato. Environmental Monitoring and Assessment, 193, 1–14. https://doi.org/10.1007/s10661-021-09494-0
Farrús, M., Anguita, J., Hernando, J., & Cerdà, R. (2007). Fusión de sistemas de reconocimiento basados en características de alto y bajo nivel. In III Congreso da Sociedade Española de Acústica Forense: actas do Congreso; 2005 oct 27-28; Santiago de Compostela, España. Santiago de Compostela: Dirección Xeral de Creación e Difusión Cultural.
Ferreira, B. M., Soares-Filho, B. S., & Quintão, F. M. (2019). The Dinamica EGO virtual machine. Science of Computer Programming, 173,3–20. https://doi.org/10.1016/j.scico.20
18.02.002
Fick, S. E., & Hijmans, R. J. (2017). WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology, 37,4302–4315. https://doi.org/10.1002/joc.5086
Flores-Villela, O., & Ochoa-Ochoa, L. M. (2020). Compilación de bases de datos de la herpetofauna mexicana. Museo de Zoología “Alfonso L. Herrera”, Facultad de Ciencias, Universidad Nacional Autónoma de México. Ciudad de México.
GBIF.org. (2020a). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
bbt2tq
GBIF.org. (2020b). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
j9evdt
GBIF.org. (2020c). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
z8nbtw
GBIF.org. (2020d). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
x4qxcx
GBIF.org. (2020e). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
apt9wm
GBIF.org. (2020f). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
bugsgy
GBIF.org. (2020g). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
x33mhm
GBIF.org. (2020h). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
esbshy
GBIF.org. (2020i). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
ju93kd
GBIF.org. (2020j). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
jrv9uw
GBIF.org. (2020k). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
gmjecq
GBIF.org. (2020l). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
q7kvhy
GBIF.org. (2020m). GBIF Occurrence Download Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
jjrncx
GBIF.org. (2020n). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
7hypb5
GBIF.org. (2020o). GBIF Occurrence Download Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
vncwkt
GBIF.org. (2020p). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
sbrrgp
GBIF.org. (2020q). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
t6y7vb
GBIF.org. (2020r). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
ksdpx3
GBIF.org. (2020s). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
uy2d7k
GBIF.org. (2020t). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
py7yby
GBIF.org. (2020u). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
8c8umf
GBIF.org. (2020v). GBIF Occurrence Download. Retrieved on November 11th, 2020 from: https://doi.org/10.15468/dl.
t92jw8
GBIF.org. (2020w). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
wumbut
GBIF.org. (2020x) GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.8tc8xr
GBIF.org. (2020y). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
tb25b4
GBIF.org. (2020z). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
paayuf
GBIF.org. (2020aa). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
8fzeuv
GBIF.org. (2020ab). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
vfa2qq
GBIF.org. (2020ac). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
ug5cus
GBIF.org. (2020ad). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
gpsjwg
GBIF.org. (2020ae). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
4ufzz3
GBIF.org. (2020af). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
dwvs69
GBIF.org. (2020ag). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.
upjycdg
GBIF.org. (2022ah). GBIF Occurrence Download. Retrieved on December 7th, 2020 from: https://doi.org/10.15468/dl.6tt8z6
Geary, W. L., Tulloch, A. I. T., Ritchie, E. G., Doherty, T. S., Nimmo, D. G., Maxwell, M. A. et al. (2023). Identifying historical and future global change drivers that place species recovery at risk. Global Change Biology, 29, 2953–2967. https://doi.org/10.1111/gcb.16661
González, T. M., González-Trujillo, J. D., Muñoz, A., & Armenteras, D. (2021). Differential effects of fire on the occupancy of small mammals in neotropical savanna-gallery forests. Perspectives in Ecology and Conservation, 19, 179–188. https://doi.org/10.1016/j.pecon.2021.03.005
Gotelli, N. J., & Colwell, R. K. (2011). Estimating species richness. In A. E. Magurran, & B. J. McGill (Eds.), Biological diversity: frontiers in measurement and assessment (pp. 39–54). Oxfordshire, England: Oxford University Press.
He, T., Lamont, B. B., & Pausas, J. G. (2019). Fire as a key driver of Earth’s biodiversity. Biological Reviews, 94, 1983–2010. https://doi.org/10.1111/brv.12544
Hortal, J., De Bello, F., Diniz-Filho, J. A. F., Lewinsohn, T. M., Lobo, J. M., & Ladle, R. J. (2015). Seven Shortfalls that Beset Large-Scale Knowledge of Biodiversity. Annual Review of Ecology, Evolution, and Systematics, 46, 523–549. https://doi.org/10.1146/annurev-ecolsys-112414-054400
Hu, F. S., Higuera, P. E., Duffy, P., Chipman, M. L., Rocha, A. V., Young, A. M., & Dietze, M. C. (2015). Arctic tundra fires: natural variability and responses to climate change. Frontiers in Ecology and the Environment, 13, 369–377. https://doi.org/10.1890/150063
IUCN (International Union for Conservation of Nature). 2001. IUCN Red List categories and criteria: Version 3.1. Prepared by IUCN Species Survival Commission. World Conservation Union, Gland, Switzerland and Cambridge, United Kingdom.
IUCN (International Union for Conservation of Nature). (2012). IUCN Red List Categories and Criteria: Version 3.1. (2nd ed.). IUCN, Gland, Switzerland.
IUCN (International Union for Conservation of Nature). (2016). Sciurus oculatus. The IUCN Red List of Threatened Species. Retrieved in 2022 from: e.T20017A22246721. https://dx.doi.org/10.2305/IUCN.UK.2016-2.RLTS.T20017A22246721.en
IUCN (International Union for Conservation of Nature). (2017). Sorex saussurei. The IUCN Red List of Threatened Species. Retrieved in 2022 from: e.T41416A22317311. https://dx.doi.org/10.2305/IUCN.UK.2017-2.RLTS.T41416A22317311.en
Jain, A., Nandakumar, K., Ross, A. (2005). Score normalization in multimodal biometric systems. Pattern Recognition, 38, 2270–2285. https://doi.org/10.1016/j.patcog.2005.01.012
Joppa, L. N., Roberts, D. L., Myers, N., & Pimm, S. L. (2011). Biodiversity hotspots house most undiscovered plant species. Proceedings of the National Academy of Sciences, 108, 13171–13176. https://doi.org/10.1073/pnas.1109389108
Kass, J. M., Vilela, B., Aiello-Lammens, M. E., Muscarella, R., Merow, C., & Anderson, R. P. (2017). Wallace: A flexible platform for reproducible modeling of species niches and distributions built for community expansion. Methods in Ecology and Evolution, 9, 1151–1156. https://doi.org/10.1111/2041-210X.12945
Kelly, L. T., Giljohann, K. M., Duane, A., Aquilué, N., Archibald, S., Batllori, E. et al. (2020). Fire and biodiversity in the Anthropocene. Science, 370, 1–10. https://doi.org/10.1126/science.abb0355
Kindt, R., & Coe, R. (2005). Tree diversity analysis. A manual and software for common statistical methods for ecological and biodiversity studies. Nairobi, Kenya: World Agroforestry Centre (ICRAF).
Leyte-Manrique, A., Mata-Silva, V., Baez-Montes, O., Fucsko, L. A., DeSantis, D. L., Garcia-Padilla, E. et al. (2022). The herpetofauna of Guanajuato, Mexico: composition, distribution, and conservation status. Amphibian & Reptile Conservation, 16, 133–180.
Lomolino, M. V. (2004). Conservation biogeography. In M. V. Lomolino, & L. R. Heaney (Eds.), Frontiers of biogeography: new directions in the geography of nature (pp. 293–296). Sunderland, Massachusetts: Sinauer.
Luedtke, J. A., Chanson, J., Neam, K., Hobin, L., Maciel, A. O., Catenazzi, A. et al. (2023). Ongoing declines for the world’s amphibians in the face of emerging threats. Nature, 622, 308–314. https://doi.org/10.1038/s41586-023-06578-4
Mandeville, P. B. (2008). Tips bioestadísticos: tema 18 ¿Por qué se deben centrar las covariables en regresión lineal? Ciencia UANL, 11, 300–305.
Martínez-Torres, H., Cantú-Férnandez, M., Ramírez, M. I., & Pérez-Salicrup, D. R. (2015). Fires and fire management in the Monarch Butterfly Biosphere Reserve. In K. S. Oberhauser, K. R. Nail, & S. Altizer (Eds.), Monarchs in a changing world: biology and conservation of an iconic butterfly (pp. 179–189). Ithaca, New York: Cornell University Press.
Montgomery, D., & Peck, E. A. (1992). Introduction to linear regression analysis. Second Edition. Hoboken, Nueva Jersey: John Wiley & Sons Ltd.
Morrone, J. J., Escalante, T., & Rodríguez-Tapia, G. (2017). Mexican biogeographic provinces: map and shapefiles. Zootaxa, 4277, 277–279. https://doi.org/10.11646/zootaxa.
4277.2.8
Murguía-Romero, M., & Villaseñor, J. L. (2000). Estimating the quality of the records used in quantitative biogeography with presence-absence matrices. Annales Botanici Fennici, 37, 289–296.
Naimi, B., Hamm, N. A. S., Groen, T. A., Skidmore, A. K., & Toxopeus, A. G. (2014). Where is positional uncertainty a problem for species distribution modelling? Ecography, 37, 191–203. https://doi.org/10.1111/j.1600-0587.2013.00205.x
Nasa Earth Data Cloud. (2020). Datos de incendios activos. NASA Official: Cerese Alber Retrieved on November 3rd, 2021 from: https://earthdata.nasa.gov/earth-observation-data/near-real-
time/firms/activefiredata
Oliveira, U., Paglia, A. P., Brescovit, A. D., de Carvalho, C. J. B., Silva, D. P., Rezende, D. T. et al. (2016). The strong influence of collection bias on biodiversity knowledge shortfalls of Brazilian terrestrial biodiversity. Diversity and Distributions, 22,1232–1244. https://doi.org/10.1111/ddi.12489
Osorio-Olvera, L., Lira-Noriega, A., Soberón, J., Peterson, A. T., Falconi, M., Contreras-Díaz, R. G. et al. (2020). NTBOX: an R package with graphical user interface for modelling and evaluating multidimensional ecological niches. Methods in Ecology and Evolution, 11, 1199-1206. https://doi.org/10.1111/2041-210X.13452
Pearson, R. G., Raxworthy, C. J., Nakamura, M., & Townsend, A. (2007). Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. Journal of Biogeography, 34, 102–117. https://doi.org/10.1111/j.1365-2699.2006.01594.x
Peterson, A. T., Papeş, M., & Soberón, J. (2008). Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecological Modelling, 213,63–72. https://doi.org/10.1016/j.ecolmodel.2007.11.008
Pilliod, D. S., Bury, R. B., Hyde, E. J., Pearl, C. A., & Corn, P. S. (2003). Fire and amphibians in North America. Forest Ecology and Management, 178, 163–181. https://doi.org/10.1016/S0378-1127(03)00060-4
Pyne, S. J. (2021). The Pyrocene: how we created an Age of Fire, and what happens next. Oakland, California: University of California Press.
QGIS Development Team. (2020). QGIS Geographic Information System. Open Source Geospatial Foundation Project. Beaverton, Oregon. Retrieved on March 2nd, 2021 from: https://qgis.org
Riddle, B. R., Ladle, R. J., Lourie, S. A., & Whittaker, R. J. (2011). Basic Biogeography: estimating biodiversity and mapping nature. In R. J. Ladle, & R. J. Whittaker (Eds.), Conservation Biogeography (pp. 45–92). Hoboken, New Jersey: Wiley-Blackwell. https://doi.org/10.1002/9781444390001.ch4
Rocchini, D., Hortal, J., Lengyel, S., Lobo, J. M., Jiménez-Valverde, A., Ricotta, C. et al. (2011). Accounting for uncertainty when mapping species distributions: the need for maps of ignorance. Progress in Physical Geography: Earth and Environment, 35, 211–226. https://doi.org/10.
1177/0309133311399491
Rodrigues, J. F. M., Villalobos, F., Iverson, J. B., & Diniz-Filho, J. A. F. (2019). Climatic niche evolution in turtles is characterized by phylogenetic conservatism for both aquatic and terrestrial species. Journal of Evolutionary Biology, 32,66–75. https://doi.org/10.1111/jeb.13395
Roth, D., Moreno-Sánchez, R., Torres-Rojo, J. M., & Moreno-Sánchez, F. (2016). Estimation of human induced disturbance of the environment associated with 2002, 2008 and 2013 land use/cover patterns in Mexico. Applied Geography, 66, 22–34. https://doi.org/10.1016/j.apgeog.2015.11.009
RStudio Team. (2020). RStudio: integrated development for R. RStudio, PBC, Boston, Massachusetts. Retrieved in 2020 from: http://www.rstudio.com/
Salazar, D. N., Farfán-Gutiérrez, M., & Arellano-Reyes, M. A. (2019). Cartografía de la severidad de los incendios forestales (2017, 2018, 2019) en el estado de Guanajuato empleando imágenes Sentinel-2. Jóvenes en la Ciencia, 5, 1-6.
Sánchez O., Charre-Medellín, J. F., Téllez-Girón, G., Báez-Montes, Ó., & Magaña-Cota, G. (2016). Mamíferos silvestres de Guanajuato: actualización taxonómica y diagnóstico de conservación. In M. Briones-Salas, Y. Hortelano-Moncada, G. Magaña-Cota, G. Sánchez-Rojas, & J. E. Sosa-Escalante, (Eds.), Riqueza y conservación de los mamíferos en México a nivel estatal (pp. 243–280). Mexico City: Instituto de Biología, UNAM/ Asociación Mexicana de Mastozoología/ Universidad de Guanajuato.
Santos-Barrera, G., & Flores-Villela, O. (2004). Lithobates megapoda. IUCN Red List of Threatened Species. Versión 2013.2. Cambridge, United Kingdom. Retrieved on April 1st, 2021 from: www.iucnredlist.org
SMAOT (Secretaría de Medio Ambiente y Ordenamiento Territorial). (2022). Áreas Naturales Protegidas. Gobierno del Estado de Guanajuato, Guanajuato. Retrieved on October 26th, 2022 from: https://smaot.guanajuato.gob.mx/sitio/areas-naturales-protegidas
Soberón, J., & Peterson, A. T. (2005). Interpretation of Models of Fundamental Ecological Niches and Species’ Distributional Areas. Biodiversity Informatics, 2, 1–10. https://doi.org/10.17161/bi.v2i0.4
Shlisky, A., Waugh, J., González, P., González, M., Manta, M., Santoso, H. et al. (2007). Fire, ecosystems and people: threats and strategies for global biodiversity conservation. Global Fire Initiative Technical Report 2007-2; Arlington, VA: The Nature Conservancy.
Tessarolo, G., Ladle, R. J., Lobo, J. M., Rangel, T. F. & Hortal, J. (2021), Using maps of biogeographical ignorance to reveal the uncertainty in distributional data hidden in species distribution models. Ecography, 44, 1743–1755. https://doi.org/10.1111/ecog.05793
Troudet, J., Grandcolas, P., Blin, A., Vignes-Lebbe, R., & Legendre, F. (2017). Taxonomic bias in biodiversity data and societal preferences. Scientific Reports, 7,1–14. https://doi.org/10.1038/s41598-017-09084-6
Tyukavina, A., Potapov, P., Hansen, M. C., Pickens, A. H., Stehman, S. V., Turubanova, S. et al. (2022). Global trends
of forest loss due to fire from 2001 to 2019. Frontiers
in Remote Sensing, 3,1–20. https://doi.org/10.3389/frsen.
2022.825190
United States Geological Survey (USGS). (2021). Archivo EROS de USGS-Elevación digital- Elevación global de 30 segundos de arco (GTOPO30). Retrieved on June 18th, 2021 from: https://www.usgs.gov/centers/eros/science/usgs-eros-archive-digital-elevation-global-30-arc-second-elevation-gtopo30
Vergara-Asenjo, G., Alfaro, F. M., & Pizarro-Araya, J. (2023) Linnean and Wallacean shortfalls in the knowledge of arthropod species in Chile: Challenges and implications for regional conservation. Biological Conservation, 281, 110027. https://doi.org/10.1016/j.biocon.2023.110027
Warren, D. L., & Seifert, S. N. (2011). Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecological Applications, 21,335–342. https://doi.org/10.1890/10-1171.1
Zamudio, S. (2012). Diversidad de ecosistemas del estado de Guanajuato. In Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (Conabio)/ Instituto de Ecología del estado de Guanajuato (IEE) (Eds.), La biodiversidad de Guanajuato: estudio de estado. II (pp. 19–55). Mexico City: Conabio/ IEE.
Monitoreo poblacional y estado de conservación de la ranita del Pehuenche (Alsodes pehuenche) en el valle Pehuenche, Mendoza, Argentina
Gabriela Diaz a, b, *, Vanesa Pellegrini-Piccini a, Liliana Moreno d, Martín Palma c, e, Vanesa Bentancourt c y Valeria Corbalán f
a Universidad Nacional de Cuyo-Sede Malargüe, Campus Educativo Municipal, Facultad de Ciencias Exactas y Naturales, Rosario Vera Peñaloza y Beltrán, 5613 Malargüe, Mendoza, Argentina
b Universidad Nacional de Cuyo, Instituto de Ingeniería y Ciencias Aplicadas a la Industria-CONICET, Facultad de Ciencias Aplicadas a la Industria, Bernardo de Yrigoyen Núm. 375, 5600 San Rafael, Mendoza, Argentina
c Instituto de Educación Física Núm. 9-016 “Jorge E. Coll” Dirección General de Escuelas-Sede Malargüe, Tecnicatura en Conservación de la Naturaleza, Campus Educativo Municipal, Rosario Vera Peñaloza y Beltrán, 5613 Malargüe, Mendoza, Argentina
d Universidad Nacional de San Luis, Facultad de Química Bioquímica y Farmacia, Ejército de los Andes Núm. 950, 5700 San Luis, Argentina
e Ministerio de Ambiente y Desarrollo Sustentable de la Provincia de Mendoza, Dirección de Recursos Naturales Renovables, Delegación Malargüe, San Martín Norte Núm. 352, 5613 Malargüe, Mendoza, Argentina
f Instituto Argentino de Investigaciones de Zonas Áridas (CCT Mendoza-CONICET), Av. Ruiz Leal s/n, Parque Gral. San Martín, 5500 Mendoza, Argentina
*Autor para correspondencia: gdiaz@infoar.net (G. Diaz)
Recibido: 02 octubre 2023; aceptado: 15 agosto 2024
Resumen
La ranita del Pehuenche, Alsodes pehuenche, es endémica de los Andes centrales de Argentina y Chile, ha sido categorizada en peligro crítico por la UICN y entre sus amenazas se encuentran la ruta internacional que atraviesa los arroyos que habita, la presencia del hongo quitridio, los salmónidos exóticos invasores, el ganado y el cambio climático. El objetivo de este trabajo fue evaluar el estado actual de conservación de A. pehuenche en el valle Pehuenche para conocer tendencias poblacionales, el impacto de las amenazas y futuras acciones de manejo. Se realizaron 14 salidas de campo durante 3 temporadas (2021-2023) y se muestrearon 12 arroyos usando la técnica de encuentro visual nocturno. Se delimitaron y nombraron 7 subpoblaciones: Nacientes, del Límite, Pichintur, Rial Rojas, Nueva, Campanaria y Cajón Largo. Los resultados muestran conteos de adultos (5.82 en 200 m2 y 13.64 por hora) y de larvas (6.24 en 200 m2 y 17.76 por hora). Éstos no variaron significativamente entre temporadas, pero fueron mayores en enero y febrero. Con base en la conectividad y las amenazas, los índices del estado de conservación permiten priorizar las subpoblaciones como unidades de conservación, de las cuales la del Límite requiere esfuerzos más urgentes.
Palabras clave: Especie amenazada; Encuentro visual; Conectividad; Subpoblaciones; Priorización de conservación
© 2024 Universidad Nacional Autónoma de México, Instituto de Biología. Este es un artículo Open Access bajo la licencia CC BY-NC-ND
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Population monitoring and conservation status of the Pehuenche frog (Alsodes pehuenche) in the valle Pehuenche, Mendoza, Argentina
Abstract
The Pehuenche spiny-chest frog, Alsodes pehuenche, is endemic to the Central Andes of Argentina and Chile. It has been categorized as critically endangered by the IUCN and its threats include the international road that crosses the streams inhabited by the species, the presence of the chytrid fungus, invasive exotic salmonids, livestock, and climate change. The objective of this work was to evaluate the current conservation status of A. pehuenche in the Pehuenche Valley as a basis for understanding population trends, the impact of threats, and future management actions. Fourteen field trips were conducted during 3 seasons (2021-2023) and 12 streams were sampled using the nocturnal visual encounter technique. Seven subpopulations were delimited and named: Nacientes, del Límite, Pichintur, Rial Rojas, Nueva, Campanaria, and Cajón Largo. The results show counts of adults (5.82 in 200 m2 and 13.64 per hour) and larvae (6.24 in 200 m2 and 17.76 per hour). These did not vary significantly between seasons but were higher in January and February. According to connectivity and threats, the conservation status indices allow us to prioritize the subpopulations as conservation units, with del Límite being the one that requires the most urgent efforts.
Keywords: Threatened species; Visual encounter; Connectivity; Subpopulations; Conservation prioritization
Introducción
Los anfibios son el grupo de vertebrados más amenazados de nuestro planeta, varias son las causas responsables de la disminución de sus poblaciones (Grant et al., 2020; Green et al., 2020; Luedtke et al., 2023). Más de 45% de la diversidad de anfibios del mundo se distribuyen en el Neotrópico (Kacoliris et al., 2022). Alrededor de 25% de las especies de Argentina son endémicas (Vaira et al., 2017), 37% de ellas en Argentina y Chile se encuentran en disminución, mientras que del 22% de las especies no se conoce su tendencia poblacional (Kacoliris et al., 2022). Las amenazas más importantes con las que se asocian la disminución poblacional o extinciones locales son los depredadores invasores, enfermedades emergentes y la ganadería (Kacoliris et al., 2022; Velasco et al., 2016).
La ranita del Pehuenche, Alsodes pehuenche, fue descrita por Cei (1976). Luego de estudios citogenéticos realizados entre 1983 y 2003 (Cuevas y Formas, 2003), se renueva el interés sobre la especie en Argentina debido a las obras viales sobre la ruta internacional ARG145 – CH115, cuya pavimentación desvió el curso de 5 afluentes del arroyo Pehuenche con presencia de la especie (Corbalán et al., 2010). La ranita del Pehuenche es una especie endémica y su distribución está restringida a los Andes centrales de Argentina y Chile (Corbalán et al., 2010, 2023; Correa et al., 2013, 2018, 2020). Habita arroyos de montaña en ecosistemas de vegas o mallines entre 2,150 y 2,825 m snm (Corbalán et al., 2023). Los arroyos poseen lechos pedregosos y una fina capa de sedimentos. La especie presenta dimorfismo sexual y como otras especies de Alsodes, tiene larvas de desarrollo prolongado con juveniles y adultos de hábitos acuáticos (Cei, 1976, 1980; Herrera y Velázquez, 2016a; Úbeda, 2021). Las larvas son de gran tamaño y pasan al menos 4 años en los cuerpos de agua permanentes hasta completar el ciclo larval (Corbalán et al., 2014). Se ha reportado la puesta de huevos ocultos bajo rocas o en oquedades en las márgenes de los arroyos (Corbalán et al., 2014; Piñeiro et al., 2020). Estas cavidades son utilizadas también como refugio por los adultos (Cei, 1980; Correa et al., 2013; Herrera y Velázquez, 2016b).
Los datos de conteos disponibles corresponden a muestreos de tramos cortos de arroyos en Argentina y Chile (Corbalán et al., 2010, 2023; Correa et al., 2013). La coexistencia de individuos en una cavidad durante el día sugiere densidades elevadas en una categoría de microhábitat (Correa et al., 2013). Han ocurrido eventos de mortalidad y se han reportado nuevas poblaciones (Corbalán et al., 2023; Correa et al., 2018), lo cual hace necesaria una evaluación actualizada de su estatus de conservación y tendencia poblacional.
Las evaluaciones del estado de conservación de la ranita del Pehuenche la han colocado en la categoría más alta de amenaza: “en peligro” por la Asociación Herpetológica Argentina (Vaira et al., 2012), cuarta en orden de prioridad entre los 58 anfibios evaluados de Chile (Vidal et al., 2024) y “en peligro crítico” por la Unión Internacional para la Conservación de la Naturaleza (IUCN, 2019), según los criterios B1ab basados en su extensión de presencia estimada y su disminución continua estimada, provocada principalmente por la pavimentación de la ruta. El área de ocupación de la especie (AOO sensu UICN) estimada actualmente es de 4.84 km2 y su extensión de presencia (EOO sensu UICN) 497.9 km2 (Corbalán et al., 2023). En cuanto a las amenazas consideradas en la categorización de IUCN (2019), se enumeran el desvío de los cursos de agua por la construcción de la ruta, el impacto del ganado, el cambio climático, la presencia del hongo quitridio (Batrachochytrium dendrobatidis) y la depredación por salmónidos exóticos invasores tales como la trucha arcoíris (Oncorhynchus mykiss)y trucha marrón (Salmo trutta) (Corbalán et al., 2023; Ghirardi et al., 2014; Zarco et al., 2020).
Si bien los avances en el conocimiento de la distribución de A. pehuenche han sido importantes en los últimos años, aún se desconocen aspectos básicos de la ecología, sistemática, reproducción, comportamiento y estrategias ecofisiológicas de esta especie en los humedales de altura. Por tratarse de una especie endémica y amenazada, la historia de vida y demografía son fundamentales para la evaluación del estatus de conservación (Luja et al., 2015). Los programas de seguimiento de poblaciones son necesarios para identificar y detectar disminuciones que amenacen la persistencia de poblaciones y deben llevarse a cabo en un marco de gestión adaptativa que permita realizar monitoreos que maximicen la detección y minimicen el esfuerzo (Pollock, 2006, Yoccoz et al., 2001). A su vez, la definición de subpoblaciones es una herramienta útil que permite definir su estado actual y priorizar acciones de conservación (Velasco, 2018).
El objetivo de este trabajo fue evaluar el estado de conservación actual de A. pehuenche en el valle Pehuenche. Esta información es fundamental para estimar tendencias poblacionales a largo plazo, evaluar el impacto de las amenazas y el éxito de futuras acciones de manejo. En este trabajo se muestran los primeros datos del programa de monitoreo iniciado en 2021 en el valle Pehuenche. A partir del mismo, se definen y delimitan subpoblaciones como unidades de conservación sobre las que se deben priorizar las acciones.
Materiales y métodos
El área de estudio corresponde al valle Pehuenche, en el lado argentino de la zona limítrofe entre Argentina y Chile; forma parte de los Andes centrales, en el departamento de Malargüe, suroeste de la provincia de Mendoza. Las precipitaciones anuales son de 400 a 600 mm (Rivera et al., 2018) y están influidas por los vientos provenientes del Pacífico sur, creando un gradiente de precipitación y humedad de oeste a este con fuertes nevadas en invierno (Garreaud et al., 2009). Durante la primavera-verano, el deshielo alimenta los humedales, denominados localmente vegas o mallines, donde se asientan familias con su ganado en los puestos, denominados reales o riales. El área es considerada un corredor ecológico y cultural trashumante de gran importancia (Llano et al., 2021).
La cuenca del arroyo Chico en Argentina incluye las subcuencas donde se distribuye A. pehuenche: arroyos Pehuenche y Callao (Corbalán et al., 2023). El arroyo Chico es afluente del río Grande en el departamento de Malargüe, provincia de Mendoza. En Chile, el área de distribución de la especie se ubica en la cuenca del río Maule, con 2 subpoblaciones posiblemente aisaldas: laguna del Maule y Lo Aguirre (Correa et al., 2013, 2018).
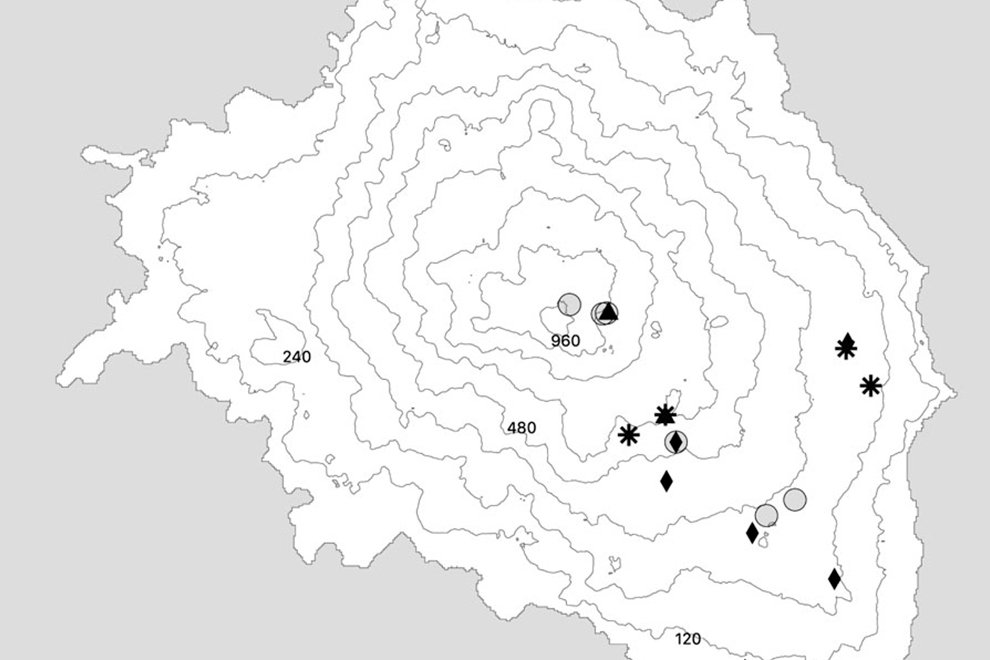
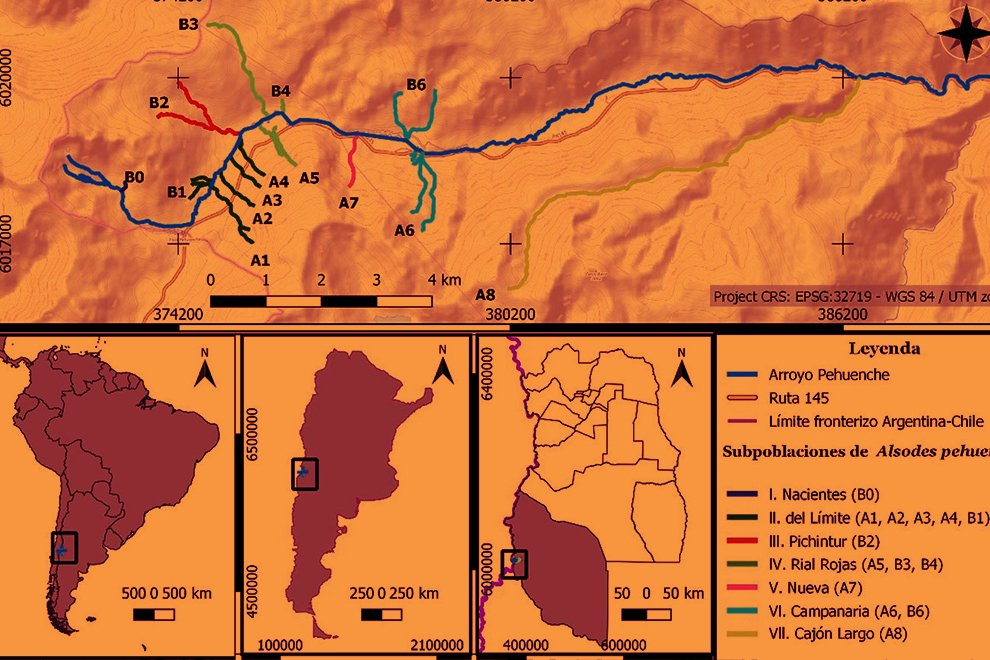
Durante 3 temporadas se muestrearon 14 arroyos tributarios del arroyo Pehuenche (fig. 1). Doce de los 14 arroyos fueron muestreados exhaustivamente y en los 2 restantes (B0 y A8), solo se constató la presencia de la especie. La especie fue observada en la desembocadura de los arroyos de primer orden en el arroyo Pehuenche.
Siguiendo a Corbalán et al. (2010) y Prado et al. (2019), los arroyos de primer orden que desembocan en el arroyo Pehuenche se distinguen, según su ubicación respecto a este último, en arroyos A que se ubican hacia el sur y son atravesados por la ruta Núm. 145, mientras que los arroyos B se ubican al norte del arroyo Pehuenche (fig. 1). A los arroyos con bifurcaciones y cursos de agua paralelos a menos de 100 m de distancia se los denominó con el mismo nombre. Durante el estudio, se tomaron datos de temperatura del agua con termómetro de mercurio, así como pH, oxígeno disuelto y conductividad con sonda multiparamétrica Lutron WA-2017SD, y de temperatura ambiente mediante la aplicación yr (Jensen et al., 2007). La conductividad fue baja, entre 0 y 181.56 μS/cm (n = 65), el valor medio del pH estuvo cercano a la neutralidad 6.41 ± 1.2, variando entre 3.29 y 8.25 (n = 77) y la concentración de oxígeno disuelto varió entre 1.3 y 15.8 mg/l (n = 16). La temperatura ambiente durante los muestreos osciló entre 2 y 21 ºC (n = 89), y la temperatura del agua entre 2 y 17.6 ºC (n = 75).
Se realizaron un total de 14 salidas de campo con una duración de 2 a 4 días/noches (tabla 1). Las 14 salidas se distribuyeron en 3 temporadas; cada una se inicia con el deshielo (octubre-noviembre-diciembre) y finaliza con la caída de las primeras nevadas (marzo-abril). En enero de 2021 se iniciaron muestreos preliminares diurnos y nocturnos consistentes en encuentros visuales sin manipulación de individuos. Los muestreos nocturnos iniciaron luego de la puesta del sol que coincide con el pico de actividad de los individuos adultos. Se optó por continuar con los muestreos nocturnos debido a que no se obtuvieron diferencias significativas en la cantidad de larvas entre el día y la noche (W = 191, p = 0.859).

Figura 1. Arroyos muestreados en la cuenca del valle Pehuenche, distribuidos en 7 subpoblaciones locales: I. Nacientes, II. del Límite, III. Pichintur, IV. Rial Rojas, V. Nueva, VI. Campanaria, VII. Cajón Largo. Los arroyos de cada subpoblación se indican con el mismo color.
Se contabilizaron adultos, juveniles y larvas usando la técnica de encuentro visual (Crump y Scott, 1994). Los transectos fueron de rumbo variable siguiendo el curso del arroyo y el ancho fue de 1 m a cada lado del arroyo. Cada transecto se realizó con 2 a 5 observadores. Siguiendo las recomendaciones de Pereyra et al. (2021), al menos un observador tenía experiencia previa y las salidas se programaron en días con condiciones meteorológicas similares: días sin lluvias, mayormente despejados y con vientos leves a moderados.
Para los estadios maduros se registró sexo (macho, hembra e indeterminado). Se registró punto GPS y hora de inicio y de finalización del transecto. Se calcularon las abundancias relativas como individuos/área e individuos/tiempo. El área de los arroyos se consideró como el tramo muestreado en metros multiplicado por 2 m de ancho. Los individuos contabilizados estuvieron a no más de 1 m de distancia del curso del arroyo, cuando éste está claramente definido, y solo se observaron más dispersos en las zonas húmedas de vegas. Para el cálculo de individuos/tiempo se usó el tiempo en minutos de la duración del muestreo (Pereyra et al., 2021).
Adicionalmente, se realizaron muestreos en transectos de 800 m de longitud en los márgenes de la calzada: 10 en la temporada estival de 2021 y 5 en las temporadas siguientes. Este registro se realizó ya que en enero de 2018 se observaron individuos deshidratados o muertos, adyacentes a un cordón o bordillo de 20 cm de altura y 15 cm de ancho.
Se definieron subpoblaciones, siguiendo a Velasco (2018), formadas por un grupo de arroyos que contienen individuos de A. pehuenche, con hábitat adecuado (sensu Corbalán et al., 2010; Correa et al., 2013) y cuya distancia entre ellos no fue mayor a 500 m lineales. De este modo, los 14 arroyos iniciales fueron agrupados en 7 subpoblaciones (fig. 1). Los nombres de las subpoblaciones se relacionan con referencias a las características del paisaje donde se encuentran, excepto la subpoblación denominada Nueva. Se calculó el índice de conectividad (IC) entre subpoblaciones de acuerdo con la fórmula de Lin (2009), modificada por Velasco (2018): IC = A1*(1/D1*T1) + A2*(1/D2*T2), donde Ai es el área (= tamaño poblacional) de la población local vecina hacia el lado i, Di es la distancia a la población local vecina hacia el lado i y Ti la presencia de truchas en hábitat intermedios (presencia multiplica por 2, ausencia multiplica por 1). Para estimar el área correspondiente a cada subpoblación, se usaron las coordenadas GPS de los muestreos de campo y en el caso de las 2 subpoblaciones donde solo se constató presencia, se estimó tamaño del área en Google Earth Pro.
Tabla 1
Temporadas y subpoblaciones en las que se realizaron los muestreos de Alsodes pehuenche. Se indican la cantidad de muestreos realizados en la ruta y los nombres de los arroyos (tipos A, B; P: arroyo Pehuenche) muestreados por subpoblación.
| Temporada | Mes | Quincena primera (1) segunda (2) | Ruta | Subpoblaciones | ||||||
| I | II | III | IV | V | VI | VII | ||||
| 1 (2021) | enero | 1 | 2 | B0 | A1, A2 | |||||
| enero | 2 | 6 | A1, A2, A3, A4 | A5 | A7 | |||||
| febrero | 2 | 1 | A1, A2 | B2 | B3 | |||||
| abril | 2 | 1 | A1, A2 | A5 | A8 | |||||
| 2 (2021-2022) | noviembre | 2 | 1 | A1, B1 | B2 | A5 | A7 | |||
| febrero | 1 | A2 | B3 | |||||||
| marzo | 1 | A1 | B2 | |||||||
| marzo | 2 | B1 | B3 | |||||||
| 3 (2022-2023) | noviembre | 2 | 1 | A5 | ||||||
| diciembre* | 1 | 1 | A1, A3, B1 | B2, B2 | A5-B3 | A6 | A7 | |||
| enero | 1 | 1 | A2, P | A5, B3, P | A6, P | A7, P | ||||
| febrero | 2 | 1 | A1, P | A7, P | ||||||
| marzo | 2 | A3, P | B4, P | B6, P |
Las amenazas que se registraron durante los conteos se consideraron de manera cualitativa, en una escala de 0 a 3, de acuerdo con la intensidad de la amenaza (Velasco, 2018). En la tabla 2 se muestran las amenazas consideradas: 1) la presencia de salmónidos que fueron registrados por encuentro visual; 2) la sequía observada de arroyos que podría ser reversible según la época del monitoreo y cárcavas permanentes cuya profundidad va en aumento; 3) la presencia de ganado, principalmente vacuno y caprino, el primero es el que podría causar mayor impacto debido al mayor uso de las vegas; 4) mortalidad de individuos por causas indeterminadas; 5) la ruta, considerada como una barrera para la dispersión, ya sea porque las alcantarillas no fueron diseñadas como pasos de fauna o porque la altura del cordón o bordillo construido sobre la misma es muy alto para el libre tránsito de ranas juveniles y adultas. Y a pesar de que la ruta representa una barrera, la construcción posterior de rampas, con el objetivo de mitigar este impacto, se considera una acción concreta de conservación. Se registró, además, la presencia de depredadores no acuáticos potenciales y de otras especies de anfibios, pero ninguno de estos casos se considera como amenaza, aunque podrían representar interacciones interespecíficas negativas.
El estado de conservación (EC) relativo de cada subpoblación se obtuvo a partir de la sumatoria de los valores asignados al índice de conectividad y las amenazas (Velasco, 2018). Los valores fueron estandarizados (ECE = EC subpoblación – promedio de todos los EC/desviación estándar de todos los EC) y el valor resultante se multiplicó por -1. Así, los valores negativos de ECE quedaron relacionados con situaciones menos favorables. Se establecieron 2 categorías de prioridades de conservación: baja a media (valores positivos de ECE) y alta (valores negativos de ECE). Los valores altos son los que requieren medidas más urgentes de manejo (Velasco, 2018).
Tabla 2
Variables consideradas para evaluar estado de conservación de las subpoblaciones de Alsodes pehuenche en el valle Pehuenche. IC: Índice de conectividad. Valores 0 a 3 son los que se le asignan a cada variable.
| Variables | 0 | 1 | 2 | 3 |
| Área (m2) | 7,500-10,000 | 5,000-7,500 | 2,500-5,000 | 1,000-2,500 |
| IC1 | 7.51-10 | 5.1-7.5 | 2.51-5 | 0-2.5 |
| Amenazas | ||||
| Efecto especies exóticas: salmónidos | sin | presencia registrada | presencia registrada en más de un conteo | presencia registrada y presencia de anfibio/s con signo de depredación |
| Fragmentación por la ruta | Sin ruta | Sin cordón o bordillo | Con cordón o bordillo y con rampas | Con cordón o bordillo y sin rampas |
| Pérdida de hábitat por sequía/ cárcavas | Sin evidencias de sequía ni cárcavas | Algún arroyo parcialmente seco | Algún arroyo parcialmente seco y/o con presencia cárcavas | Algún arroyo completamente seco y/o presencia de individuos muertos por sequía |
| Efecto del ganado | Sin ganado | Con caprinos | Con vacunos | Con vacunos y cabras |
| Indeterminada | Sin registro de individuos muertos sin causa aparente | Registro de individuos con manchas en la piel | Registro de individuos con manchas y uno muerto sin causa aparente | Registro de más de un individuo muerto sin causa aparente |
1 = Índice de conectividad calculado de acuerdo con Lin (2009), modificado por Velasco (2018).
Se realizaron análisis de los conteos con R versión 4.1.3 y los análisis estadísticos consideraron 0.05 de nivel de significación de alfa. La normalidad de los datos se verificó con la prueba de Shapiro-Wilk. La cantidad de individuos (adultos, juveniles y larvas) por área y por hora no se distribuyó normalmente (W < 0.82934, p < 0.001), por lo que se usaron las pruebas estadísticas no paramétricas de Mann-Whitney-Wilcoxon y de Kruskal-Wallis para 2 grupos y más de 2 grupos, respectivamente. Se evaluó estadísticamente si hubo diferencias entre adultos y juveniles considerados en conjunto versus larvas. También se evaluaron diferencias en relación con variables temporales (temporadas, meses) y espaciales (entre arroyos A y B y entre subpoblaciones). Cuando se obtuvieron diferencias significativas con la prueba de Kruskal-Wallis, se usó el paquete conover.test versión 1.1.5 (2017), basado en Conover-Iman Test para comparaciones múltiples. En todos los casos, los resultados se expresan en mediana, el rango intercuartílico (IQR) como medida de dispersión, los valores mínimos y máximos (min-max) y el tamaño de la muestra (n).
Resultados
Los transectos muestreados alcanzaron un total de 51,292 m lineales recorridos en 181 horas con un esfuerzo de muestreo 12.35 horas/persona. De los 14 arroyos estudiados, todos tuvieron presencia de la especie y el área total fue de 0.04 km². Más de la mitad corresponde a los arroyos denominados A (56.2%).
Cuando se analizaron los conteos respecto de la cantidad de observadores, la correlación de Spearman resultó en valores de rho cercanos a 0, lo que sugiere que no hay correlación lineal (p > 0.05), tanto para individuos postmetamórficos (por área: rho = -0.0427, n = 99; por hora: rho = 0.0975, n = 99), como para larvas (por área: rho = -0.0297, n = 92; por hora rho = -0.0063,
n = 92).
A lo largo del periodo de muestreo se observaron 5.82 adultos y juveniles/área (IQR = 9.27, 0-45 min-max, n = 99) y 13.64 adultos y juveniles/hora (IQR = 30.29, 0-108 min-max, n = 99); 6.24 larvas/área (IQR = 31, 0-175 min-max, n = 92) y 17.76 larvas/hora (IQR = 65, 0-367 min-max, n = 92). La comparación entre estadios muestra menos postmetamórficos que larvas, tanto para los conteos de individuos por área (W = 12.524, p < 0.001) como por hora (W = 26.918, p < 0.001).
Tabla 3
Prioridades de conservación para las subpoblaciones de Alsodes pehuenche en el valle Pehuenche, con base en el tamaño de la población, la conectividad y las amenazas.
| Sub-población | Área (m2) | IC | Amenazas | ECE | PC1 | ||||
| Ruta | Sequía/cárcavas | Ganado | Muertes indet. | Salmónidos | |||||
| I. Nacientes | 4,044 | 8.41 | 0 | 0 | 2 | 0 | 1 | 0.88 | Baja |
| II. del Límite | 5,744 | 8.39 | 2 | 3 | 2 | 2 | 3 | -1.26 | Muy alta |
| III. Pichintur | 5,002 | 9.91 | 0 | 0 | 2 | 3 | 2 | 0.08 | Media |
| IV. Rial Rojas | 6,208 | 6.05 | 1 | 3 | 2 | 1 | 2 | -0.73 | Alta |
| V. Nueva | 2,160 | 9.61 | 1 | 0 | 2 | 3 | 0 | -0.19 | Media |
| VI. Campanaria | 5,860 | 5.26 | 1 | 0 | 1 | 3 | 3 | -0.46 | Media |
| VII. Cajón Largo | 9,550 | 6.64 | 0 | 0 | 1 | 0 | 0 | 1.69 | Muy baja |
1 PC: Prioridad de conservación calculada de acuerdo con Velasco (2018).
Las comparaciones entre las 3 temporadas no mostraron diferencias significativas para ninguno de los grupos estudiados adultos y juveniles/área (H = 1.9935, df = 2, p = 0.369), adultos y juveniles/hora (H = 1.3599, df = 2, p = 0.5066), larvas/área (H = 0.13936, df = 2, p = 0.933) y larvas por hora (H = 0.6051, df = 2, p = 0.7389). Entre meses, la diferencia no fue significativa para larvas (H = 4.5278, df = 5, p = 0.4762 por área, H = 4.0986, df = 5, p = 0.5353 por hora), en cambio para los postmetamórficos solo fue significativa para los conteos por área (H = 13.97, df = 5, p = 0.0158). Los análisis post hoc indican menores cantidades de adultos y juveniles en febrero respecto a diciembre (p = 0.0075) y enero (p = 0.0021), como también en marzo respecto a enero (p = 0.0185, fig. 2).
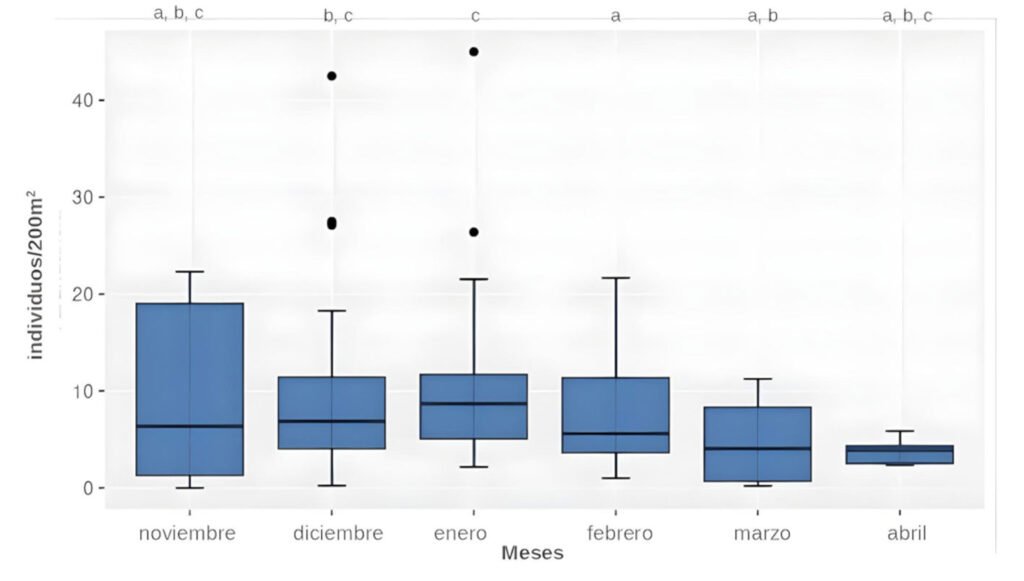
Figura 2. Variación mensual de la abundancia de adultos y juveniles (individuos/200 m2) de Alsodes pehuenche. Los meses con diferentes códigos de letras indican diferencias significativas (p < 0.05).
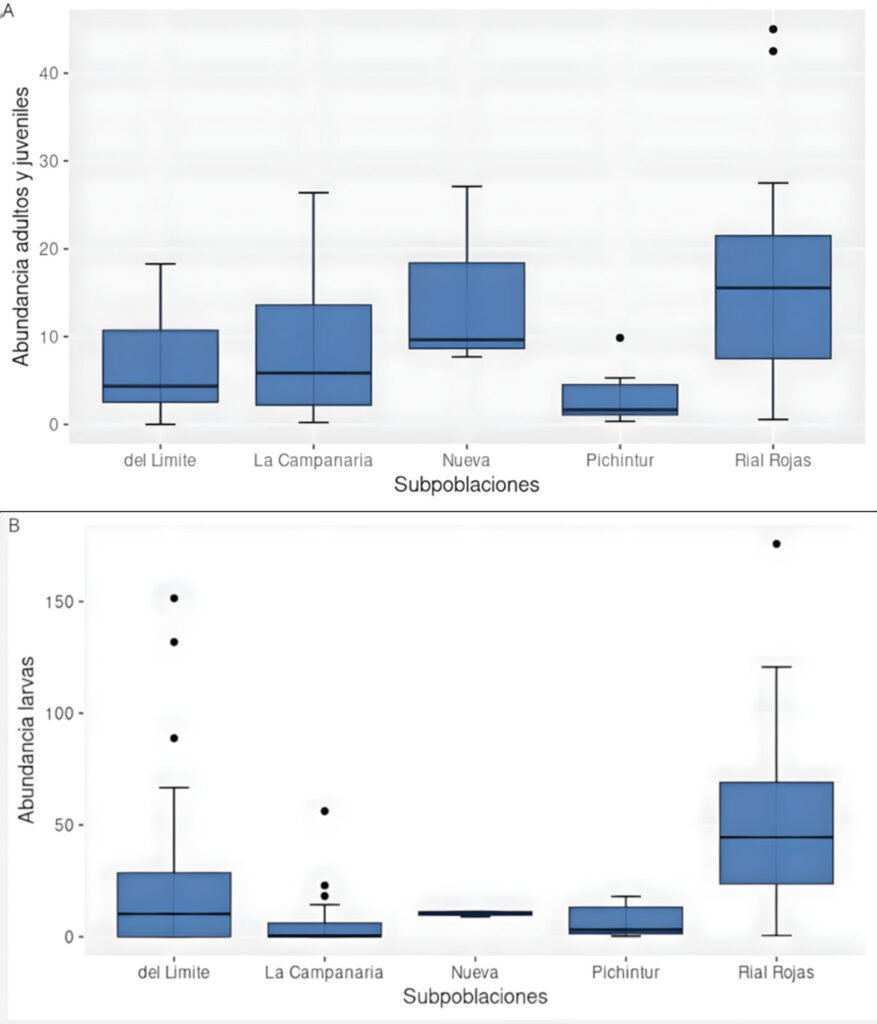
Figura 3. Abundancia de Alsodes pehuenche (individuos/200 m2) entre 5 subpoblaciones. A, Adultos y juveniles; B, larvas. Las subpoblaciones con diferentes códigos de letras indican diferencias significativas (p < 0.05).
De los 14 arroyos del valle Pehuenche con presencia de A. pehuenche, los arroyos A tienen un área de ocupación de 49% respecto de los arroyos B con 51%. Y en los 12 arroyos donde se realizaron conteos por área, éstos fueron mayores en los arroyos A que en los B, tanto para adultos y juveniles (H = 31.977, df = 2, p < 0.001), como para larvas (H = 10.42, df = 2, p = 0.0055).
Se reconocieron 7 subpoblaciones en el valle Pehuenche: Nacientes, del Límite, Pichintur, Rial Rojas, Nueva, Campanaria y Cajón Largo (fig. 1). De éstas, se tienen datos de conteo de 5 (todas, excepto Nacientes y Cajón Largo). Cuando se comparan estas 5 subpoblaciones, se encuentran diferencias significativas tanto para adultos y juveniles/área (H = 17.436, df = 4, p = 0.0016), como para larvas/área (por área H = 23.639, df = 4, p < 0.001). Se registraron más adultos y juveniles en las subpoblaciones Rial Rojas y Nueva, y más larvas en Rial Rojas (fig. 3). Sin embargo, estas diferencias no son significativas cuando se analizan los datos de postmetamórficos/hora (H = 8.8053, df = 5, p = 0.1171) y larvas/hora (H = 4.0986, df = 5, p = 0.5353).
El único lugar donde se encontraron solo larvas fue en una charca de la subpoblación del Límite. Éstas coexistían con larvas y adultos de rana de 4 ojos (Pleurodema bufoninum). Además, esta última especie ha sido observada eventualmente coexistiendo con A. pehuenche en las subpoblaciones Rial Rojas, Campanaria y del Límite. Otras especies registradas durante los muestreos, que se identifican como posibles depredadores de A. pehuenche son el zorro gris (Lycalopex gymnocercus), el chiñe o zorrino (Conepatus chinga), la gaviota capucho café (Chroicocephalus maculipennis) y el águila mora (Geranoaetus melanoleucus).
Aunque la detección de huevos no fue objeto de este estudio, dado que no se realizó búsqueda activa en oquedades ni se manipularon hembras, en noviembre 2022 se observó una masa de 20 huevos aproximadamente, flotando en una pequeña poza de la subpoblación Rial Rojas en época de deshielo. Los huevos no eran pigmentados y cada uno midió entre 6 y 7 mm de diámetro.
Los índices de conectividad calculados para cada una de las subpoblaciones se muestran en la tabla 3. El arroyo Pehuenche representa la principal conexión entre subpoblaciones. En este arroyo se encontraron individuos adultos en la desembocadura de arroyos de primer orden pertenecientes a las subpoblaciones del Límite, Rial Rojas, Nueva y Campanaria (tabla 1). Si bien Cajón Largo tiene relativamente bajo índice de conectividad, el área de esta subpoblación es la mayor de todas.
Se registraron 24 individuos muertos durante el periodo de estudio: 1) 6 individuos adultos —2 hembras, 2 machos y 2 adultos indeterminados— en el mes de enero de la temporada estival 2021 y 2 en la temporada 2022-2023 en los muestreos realizados en la ruta internacional ARG145; 2) 3 individuos —1 adulto indeterminado y 2 larvas— encontrados muertos por desecación sobre el curso de un arroyo de Rial Rojas que mostró una disminución abrupta del caudal luego del deshielo entre noviembre y diciembre 2022; 3) 4 adultos —3 machos y 1 indeterminado— encontrados con signos de depredación (patas traseras lastimadas o mutiladas) al costado o dentro del curso del arroyo donde se registra presencia de salmónidos, esto es, en las subpoblaciones del Límite y Campanaria; 4) 6 adultos —2 machos y 4 indeterminados—, 1 juvenil indeterminado y 2 larvas encontrados muertos sin causa evidente, dentro del curso del arroyo o próximo a éste.
Finalmente, los valores de ECE negativos usados como indicadores, permitieron identificar a las subpoblaciones del Límite y Rial Rojas como unidades de conservación prioritarias, de las cuales la del Límite requiere esfuerzos más urgentes.
Discusión
Corbalán et al. (2010) reportaron 355 individuos de A. pehuenche en 2,500 m lineales (equivalente a 14 individuos/200 m2) en un muestreo realizado en 2008 en arroyos de la subpoblación del Límite. Si se compara ese valor con el obtenido en este estudio para los mismos arroyos en el mismo mes en 2021 (8 individuos/200 m2), el primero es mayor, pero se encuentra dentro del intervalo de variación de este estudio. Cabe mencionar que se usaron 2 técnicas diferentes para el conteo (búsqueda activa de individuos en oquedades durante el día y encuentro visual nocturno, con 3 observadores en los 2 casos), por lo que, con la información disponible actualmente, solo la mortalidad de individuos reportada desde 2008 permite estimar una tendencia poblacional decreciente para esta subpoblación.
En 2 arroyos muy cercanos a la subpoblación del Límite en Chile, Correa et al. (2013) reportaron 20 y 24 adultos/h o 15 y 66 adultos/200 m². Estos últimos datos corresponden a muestreos nocturnos realizados en marzo de 2012, con 4 observadores. La cantidad máxima de adultos por área observados por Correa et al. (2013) es mayor al máximo obtenido en este estudio, lo cual puede deberse a un error de muestreo en solo 30 m lineales, o bien a fluctuaciones poblacionales naturales (Kissel et al., 2020). En arroyos de Rial Moreno, a 21 km al NE de la población del valle Pehuenche, Corbalán et al. (2023) reportaron 26 adultos/hora y 4 juveniles/hora en un muestreo nocturno realizado en enero de 2019, los cuales se encuentran dentro de valores obtenidos en el valle Pehuenche en este estudio.
Durante las 3 temporadas consecutivas muestreadas en este trabajo, no se detectaron cambios significativos para ningún estadio de desarrollo de A. pehuenche. Esto puede deberse a que se trata de un periodo corto en términos de monitoreo. Por lo tanto, es necesario mantener los muestreos para conocer tendencias poblacionales a largo plazo. Por otro lado, la detección de individuos por encuentro visual a lo largo de los arroyos, podría compararse con otros estudios enfocados al monitoreo poblacional de la especie para lograr establecer un óptimo en términos de esfuerzo de muestreo. Dado que estas investigaciones dependen del presupuesto disponible y de limitaciones logísticas (Joseph et al., 2006) principalmente ligadas a la elevada demanda de horas/persona en un ambiente de alta montaña, se debe buscar una estrategia que permita satisfacer tanto la demanda de datos pertinentes para la gestión, como la obtención de información ecológica de calidad (Stephens et al., 2015; Yoccoz et al., 2001).
En relación con la primera acción de conservación llevada a cabo en la población del valle Pehuenche, que consistió en la construcción de 800 m de cordón o bordillo en la ruta y 2 cámaras de infiltración para evitar que la sal vertida en la ruta llegue a los arroyos, provocó la deshidratación y muerte de 18 individuos que fueron encontrados en diciembre de 2017, y 50 en enero de 2018. Posiblemente, la construcción a posteriori de rampas entre el cordón o bordillo y la calzada en febrero de 2018, ha evitado que se vuelvan a registrar mortalidades tan abruptas. Si bien nunca pudo constatarse el uso de las rampas por parte de la especie, las mismas habrían mejorado la conexión entre las secciones superior e inferior de los arroyos. Esto demuestra la necesidad de seguimiento de las acciones de conservación que se llevan a cabo. Pero, por otro lado, si bien la ruta pudo haber provocado una disminución poblacional (Corbalán et al., 2010; IUCN, 2019), el número elevado de individuos muertos registrado en este estudio alerta sobre la necesidad de avanzar en el conocimiento de otras amenazas, como la depredación por salmónidos invasores, el cambio climático y la infección por el hongo quitridio (Batrachochytrium dendrobatidis) de los anfibios.
En este trabajo, de todos los estadios evaluados, las larvas fueron las más abundantes, las más estables en el tiempo y presentaron el mismo patrón de variación que los adultos entre arroyos A y B, y subpoblaciones (fig. 3). Al igual que lo reportado por Corbalán et al. (2010, 2023), se encontraron larvas de diferentes tamaños conviviendo en pozas, pero también se observaron en rápidos, remansos y debajo de la vegetación acuática. Estos resultados son coherentes con la estrategia de especies con desarrollo larval plurianual (Úbeda, 2021). Por otro lado, la menor cantidad de juveniles registrada respecto de adultos podría ser un problema de detectabilidad por su tamaño (Petrovana y Schmidt, 2019), podría corresponder a una segregación por microhábitats según el estadio (Gonwouo et al., 2022), o bien, ser evidencia de una dinámica poblacional particular solo en esta fase de su ciclo de vida (Kissel et al., 2020).
Los muestreos preliminares realizados de día y de noche demuestran que hay arroyos permanentes que solo tienen adultos. Los estudios a futuro podrán determinar las causas que posiblemente puedan estar asociadas con aspectos fisicoquímicos del agua, o a la dispersión de individuos. La información actual permite suponer que las rutas de dispersión de A. pehuenche con base en sus hábitos acuáticos, son los mismos arroyos. Los registros de individuos secos en la ruta, principalmente entre diciembre y enero, al igual que la mayor cantidad de adultos y juveniles registrados en estos meses (fig. 2), podrían estar indicando una mayor dispersión en esa temporada del año.
Los arroyos del valle Pehuenche son los mejores conocidos hasta el momento y el área que ocupan representa solo 6% del área de distribución de A. pehuenche (487.9 km²) reportada por Corbalán et al. (2023). De la misma manera, habiendo considerado en nuestro estudio casi todos los arroyos presentes en el valle Pehuenche, representa 0.8% del área de ocupación total para la especie (4.84 km²; Corbalán et al., 2023). El espacio sin ocupación efectiva de la especie, actualmente, es muy grande (Corbalán et al., 2023), aunque en el pasado pudo haber estado presente en un área mayor si se comparan los registros de presencia en Cajón Grande (Cei y Roig, 1965; Corbalán et al., 2023). Si se considera desde la descripción de la especie en 1965, A. pehuenche ha reducido su área de distribución histórica, lo cual puede deberse a diferentes causas. La depredación por salmónidos en la actualidad podría ser la causa de que la especie haya quedado restringida solo a las zonas altas de las cuencas. Por ello, es necesario conocer la distribución de salmónidos e identificar dónde coexisten estos peces y la rana. Donde esto ocurre en el valle Pehuenche, se registraron adultos muertos de A. pehuenche con signos de depredación. Así, los salmónidos representan una barrera para la dispersión de las ranas, reduciendo la conectividad entre subpoblaciones y dentro de ellas entre arroyos A y B (tabla 3). Como se ha demostrado para otras especies de anfibios, la consecuencia es la disminución del intercambio genético o demográfico (e.g., Kacoliris et al., 2022; Velasco, 2018). Por lo tanto, para conservar las subpoblaciones se requiere asegurar su conectividad, tanto entre las secciones inferior y superior de los arroyos A impactados por la ruta, como dentro de las subpoblaciones entre los arroyos A y B, y entre las subpoblaciones conectadas a través del arroyo Pehuenche.
Las observaciones de campo sugieren que los individuos se distribuyen de manera agrupada a lo largo de los cursos de agua y vegas, no solo las larvas en las pozas, sino también los adultos a lo largo de los arroyos. Esto debe ser evaluado en la etapa reproductiva a inicios de la temporada y al final de ésta, cuando el caudal disminuye y algunos cauces se secan total o parcialmente, donde solo quedan pozas pequeñas con agua. Si este efecto es acentuado por la disminución de cobertura permanente de nieve en la cuenca del Río Grande (Aumassanne et al., 2019), es posible que más arroyos, que todavía mantienen agua hasta el final de la temporada, se sequen como ha ocurrido en los arroyos A3 y A4 de la población del Límite, aumentando así la fragmentación del hábitat. Si A4 no se hubiera secado, las 3 subpoblaciones (del Límite, Pichintur y Rial Rojas) estarían conectadas, formando una sola subpoblación (fig. 1).
Si bien gran parte de las amenazas han sido identificadas (Corbalán et al., 2010, 2023), la mayoría no han sido cuantificadas. Con base en las observaciones realizadas durante los muestreos en este estudio, se sistematizaron las amenazas para cada arroyo y cada subpoblación en el valle Pehuenche (tablas 2, 3). Como resultado de la priorización, las subpoblaciones del Límite y Rial Rojas se identifican como prioritarias para iniciar acciones concretas de conservación. Es posible que ambas sean fragmentos de una población mayor y que hayan estado unidas cuando el arroyo intermedio no estaba seco. En la del Límite se podría iniciar la mitigación de la depredación por salmónidos y reducir los efectos de la ruta. Es necesario continuar con el seguimiento y cuantificación de las amenazas, considerar las aún no evaluadas, tales como las que pueden estar generando las actividades turísticas, el vertido de sal en la ruta, crecidas extraordinarias y otras potenciales como el tendido de líneas de alta tensión, planificada desde el Maule (Chile) al río Diamante (San Rafael, Argentina).
El avance logrado en 20 años de estudios sobre la biología y conservación de este anfibio endémico de Argentina y Chile nos brinda herramientas para sostener acciones concretas de manejo que alivien alguna de las amenazas y que aseguren la viabilidad de las poblaciones a largo plazo. Se espera que un programa de monitoreo anual permita avanzar en el conocimiento de las tendencias poblacionales a largo plazo. Las subpoblaciones del valle Pehuenche son las mejor conocidas hasta el momento y han sufrido disminuciones, fragmentaciones y también extinciones locales. La priorización de su estado de conservación brinda herramientas para implementar acciones necesarias a corto plazo.
Agradecimientos
Por la colaboración en el trabajo de campo, se agradece a estudiantes de la Tecnicatura en Conservación de la Naturaleza Sede Malargüe del IEF Núm. 9-016, especialmente a Julián Rodríguez, Armando Barros, Francisco Jofré y Pablo Lucero; también a estudiantes voluntarios y Graciela Ríos. A Karen Olate por la elaboración del mapa. Y a un revisor anónimo por sus aportes a la primera versión del manuscrito y a los dos revisores que realizaron aportes importantes para mejorar el manuscrito. La investigación se logró gracias al financiamiento del proyecto SIIP 06/M003 Resol. 3978/2022 UNCUYO y aportes personales de investigadoras y estudiantes.
Referencias
Aumassanne, C. M., Beget, M. E., Di Bella, C. M., Oricchio, P. y Gaspari, F. J. (2019). Cobertura de nieve en las cuencas de los ríos Grande y Barrancas (Argentina) y su relación con la morfometría. Revista de Investigación Agropecuaria, 45, 394–403.
Cei, J. M. (1976). Remarks on some neotropical amphibians of the genus Alsodes from southern Argentina. Atti della Società Italiana di Scienze Naturali e del Museo Civico di Storia Naturale di Milano, 117, 159–164.
Cei, J. M. (1980). Amphibians of Argentina. Monitore Zoologico Italiano, N.S. Monografia 2. Florencia: Universitá degli studi di Firenze.
Cei, J. M. y Roig, V. G. (1965). The systematic status and biology of Telmatobius montanus Lataste (Amphibia: Leptodactylidae). Copeia, 4, 421–425.
Corbalán, V., Debandi, G. y Martínez, F. (2010). Alsodes pehuenche (Anura: Cycloramphidae): past, present and future. Cuadernos de Herpetología, 24, 17–23.
Corbalán, V., Debandi, G., Martínez, F. y Úbeda, C. (2014). Prolonged larval development in the critically endangered Pehuenche’s Frog Alsodes pehuenche: implications for conservation. Amphibia–Reptilia, 35, 283–292. http://dx.doi.
org/10.1163/15685381-00002951
Corbalán V., Debandi G., Literas S., Álvarez L., Rivera J. A., Dopazo J. et al. (2023). Newly discovered sites and potential threats for the critically endangered frog, Alsodes pehuenche, in Southern South America. Herpetological Conservation and Biology, 18, 48–56.
Correa, C., Pastenes, L., Iturra, P., Calderón, P., Vásquez, D., Lam, N. et al. (2013). Confirmation of the presence of Alsodes pehuenche Cei, 1976 (Anura, Alsodidae) in Chile: morphological, chromosomal and molecular evidence. Gayana, 77, 117–123. http://dx.doi.org/10.4067/S0717-65382013000200006
Correa, C., Zepeda, P., Lagos, N., Salinas, H., Palma, R. E. y Vásquez, D. (2018). New populations of two threatened species of Alsodes (Anura, Alsodidae) reveal the scarce biogeographic knowledge of the genus in the Andes of central Chile. Zoosystematics and Evolution, 94, 349–358.
Correa, C. J. Morales, C. Schussler y J. C. Ortiz. (2020). An enigmatic population of Alsodes (Anura, Alsodidae) from the Andes of central Chile with three species-level mitochondrial lineages. Mitochondrial DNA Part A DNA Mapping, Sequencing, and Analysis, 31, 25–34. https://doi.org/10.1080/24701394.2019.1704744
Crump, M. L. y Scott, N. J. (1994). Visual encounter surveys. En W. R.Heyer, M. A. Donnelly, R. W. Mc Diarmid, L. A. C. Hayek y M. S. Foster (Eds.). Measuring and Monitoring Biological Diversity Standard Methods for Amphibians (pp. 84–92). Washington D.C.: Smithsonian Institution Press.
Cuevas, C. C. y Formas, J. R. (2003). Cytogenetic analysis of four species of the genus Alsodes (Anura: Leptodactylidae) with comments about the karyological evolution of the genus. Hereditas, 138, 138–147. https://doi.org/10.1034/j.1601-5223.2003.01677.x
Garreaud, R., Vuille, M., Compagnucci, R. y Marengo, J. (2009). Present-day south american climate. Palaeogeography, Palaeoclimatology, Palaeoecology, 281, 180–195. http://dx.
doi.org/10.1016/j.palaeo.2007.10.032
Ghirardi, R., Levy, M. G., López, J. A., Corbalán, V., Steciow, M. M. y Perotti, M. G. (2014). Endangered amphibians infected with the chytrid fungus Batrachochytrium dendrobatidis in austral temperate wetlands from Argentina. Herpetological Journal, 24, 129–133.
Gonwouo, N. L. Schäfer, M. y Tsekané, S. J. (2022). Goliath frog (Conraua goliath) abundance in relation to frog age, habitat, and human activity. Amphibian and Reptile Conservation, 16, e319.
Grant, E. H. C., Miller, D. A. y Muths, E. (2020). A synthesis of evidence of drivers of amphibian declines. Herpetologica, 76, 101–107. https://doi.org/10.1655/0018-0831-76.2.101
Green D. M., Lannoo M. J., Lesbarrères D. y Muths E. (2020). Amphibian population declines: 30 years of progress in confronting a complex problem. Herpetologica, 76, 97–100. https://doi.org/10.1655/0018-0831-76.2.97
Herrera, F. y Velásquez, N. A. (2016a). Dimorfismo sexual en Alsodes pehuenche Cei 1976 (Amphibia, Anura, Alsodidae). Boletín Chileno de Herpetología, 3, 4–6.
Herrera, F. y Velásquez, N. A. (2016b). Uso de cuevas en Alsodes pehuenche Cei 1976 (Amphibia, Anura, Alsodidae). Boletín Chileno de Herpetología, 3, 17–20.
Jensen, I. S., Haugen, V. F. y Skålin, R. (2007). Yr. Norwegian Meteorological Institute and the Norwegian Broadcasting Corporation. Recuperado el 15 de agosto, 2024 de https://www.yr.no/nb
Joseph, L. N., Field, S. A., Wilcox, C. y Possingham, H. P. (2006). Presence-absence versus abundance data for monitoring threatened species. Conservation Biology, 20, 1679–1687. https://doi.org/10.1111/j.1523-1739.2006.00529.x
Kacoliris, F. P., Berkunsky, I., Acosta, J. C., Acosta, R., Agostini, M. G., Akmentins, M. et al. (2022). Current threats faced by amphibian populations in the southern cone of South America. Journal for Nature Conservation, 69, 126254. https://doi.org/10.1016/j.jnc.2022.126254
Kissel, A. M., Tenan, S. y Muths, E. (2020). Density dependence and adult survival drive dynamics in two high elevation amphibian populations. Diversity, 12, 478.
Lin, J. P. (2009). The functional linkage index: a metric for measuring connectivity among habitat patches using least-cost distances. Journal of Conservation Planning, 5, 28–37.
Llano, C., Durán, V., Gasco, A., Reynals, E. y Zárate, M. S. (2021). Traditional puesteros´ perceptions of biodiversity in semi-arid Southern Mendoza, Argentina. Journal of Arid Environments, 192, 104553. https://doi.org/10.1016/j.ja
ridenv.2021.104553
Luedtke, J. A., Chanson, J., Neam, K., Hobin L., Maciel, A. O., Catenazzi, A. et al. (2023). Ongoing declines for the world’s amphibians in the face of emerging threats. Nature, 622, 308–314. https://doi.org/10.1038/s41586-023-06235-w
Luja, V., Rodríguez-Estrella, H. R., Schaub, M. y Schmidt, B. R. (2015). Among-population variation in monthly and annual survival of the Baja California treefrog, Pseudacris hypochondriaca curta, in desert oases of Baja California Sur, Mexico. Herpetological Conservation and Biology, 10, 112–122.
Pereyra, L. C., Etchepare, E. y Vaira, M. (2021). Manual de técnicas y protocolos para el relevamiento y estudio de anfibios de Argentina. San Salvador de Jujuy: Editorial de la Universidad Nacional de Jujuy.
Petrovana, S. O. y Schmidt, B. R. (2019). Neglected juveniles; a call for integrating all amphibian life stages in assessments of mitigation success (and how to do it). Biological Conservation, 236, 252–260. https://doi.org/10.1016/j.biocon.
2019.04.016
Piñeiro, A., Fibla, P., López, C., Velásquez, N. y Pastenes, L. (2020). Characterization of an Alsodes pehuenche breeding site in the Andes of central Chile. Herpetozoa, 33, 21–26. https://doi.org/10.3897/herpetozoa.33.e49268
Pollock, J. E. (2006). Detecting population declines over large areas with presence-absence, time-to-encounter, and count survey methods. Conservation Biology, 20, 882–892. https://doi.org/10.1111/j.1523-1739.2006.00559.x
Prado, W. S., Meriggi, J., Martínez, F. y Corbalán, V. (2019). Ampliación del área de distribución de Alsodes pehuenche en Argentina. Revista del Museo de La Plata, 4 (Suplem.), 1R–117R.
Rivera, J. A., Marianetti, G. y Hinrichs, S. (2018). Validation of CHIRPS precipitation dataset along the Central Andes of Argentina. Atmospheric Research, 213, 437–449. https:
//doi.org/10.1016/j.pce.2022.103184
Stephens, P. A., Pettorelli, N., Barlow, J., Whittingham, M. J. y Cadotte, M. W. (2015). Management by proxy? The use of indices in applied ecology. Journal of Applied Ecology, 52, 1–6. https://doi.org/10.1111/1365-2664.12405
Úbeda, C. (2021). Estrategias reproductivas, hábitats y otros aspectos ecológicos de los anfibios altoandinos en la vertiente oriental de la Cordillera de los Andes. Boletín Chileno de Herpetología, 8, 10–21.
UICN (Unión Internacional para la Conservación de la Naturaleza), Species Survival Commission Amphibian Specialist Group. (2019). Alsodes pehuenche. IUCN Red List of threatened species, 2019. https://www.iucnredlist.org
Vaira, M., Akmentins, M., Attademo, M., Baldo, D., Barrasso, D., Barrionuevo, S. et al. (2012). Categorización del estado de conservación de los anfibios de la República Argentina. Cuadernos de Herpetología, 26 (Suplem. 1), 131–159.
Vaira, M., Pereyra, L. C., Akmentins M. S. y Bielby, J. (2017). Conservation status of Amphibians of Argentina: An update and evaluation of national assessments. Amphibian y Reptile Conservation, 11, e135.
Velasco, M. A. (2018). Dinámica poblacional y conservación de la ranita del Valcheta (Pleurodema somuncurense) (Cei, 1969), Patagonia, Argentina (Tesis doctoral). Universidad Nacional de La Plata, La Plata, Buenos Aires.
Velasco, M. A., Kacoliris, F. P., Berkunsky, I., Quiroga, S. y Williams, J. D. (2016). Current distributional status of the critically endangered Valcheta frog: implications for conservation. Neotropical Biology and Conservation, 11, 110–113. http://dx.doi.org/10.4013/nbc.2016.112.08
Vidal, M. A., Henríquez, N., Torres-Díaz, C., Collado, G. y Acuña-Rodríguez, I. S. (2024). Identifying strategies for effective biodiversity preservation and species status of Chilean amphibians. Biology, 13, 169. https://doi.org/10.33
90/biology13030169
Yoccoz, N. G., Nichols, J. D. y Boulinier, T. (2001). Monitoring of biological diversity in space and time. Trends in Ecology and Evolution, 16, 446–453. https://doi.org/10.1016/S0169-
5347(01)02205-4
Zarco, A., Corbalán, V. y Debandi, G. (2020). Depredación por truchas arcoíris invasoras en la rana de pecho espinoso Pehuenche, en peligro crítico. Journal of Fish Biology, 98, 878-880. https://doi.org/10.1111/jfb.14401
Priority areas for conservation based on endemic vascular plant species and their biocultural attributes: a case study in Sinaloa, Mexico
C. Rocío Álamo-Herrera a, María Clara Arteaga a, *, Rafael Bello-Bedoy b
a Instituto Politécnico Nacional, Centro Interdisciplinario de Investigación para el Desarrollo Integral Regional, Unidad Durango, Sigma No. 119, Fracc. 20 de Noviembre II, 34234 Victoria de Durango, Durango, Mexico
b Universidad de Guadalajara, Centro Universitario de Ciencias Biológicas y Agropecuarias, Cátedras Conahcyt-Universidad de Guadalajara, Camino Ramón Padilla Sánchez No. 2100, 45200 Zapopan, Jalisco, Mexico
*Corresponding author: d1j17kk@hotmail.com (J.F. Pío-León)
Received: 20 February 2024; accepted: 02 July 2024
Abstract
Endemic vascular plants are one of the main biodiversity indicators used to propose priority conservation areas. The richness of endemic species and corrected and weighted endemism are the most frequently used criteria, while anthropogenic or biocultural factors such as ethnobotanical value or ecological vulnerability are seldom considered. This work proposes priority conservation areas for Sinaloa, Mexico, considering the richness of its endemic species, corrected and weighted endemism, as well as ethnobotanical value, protection status, and the Priority Conservation Index (PCI). The analysis was performed in a 19 × 19 km grid and included 247 records of 78 species. The areas proposed when considering only the richness of endemic species and the weighted endemism coincided with previously known areas of high biodiversity in the state, which are areas of high collection effort and low anthropogenic impact. When considering the ethnobotanical value and protection status, the areas identified included those with greater anthropogenic impact, which contained species of biocultural and economic importance. When the PCI was used, both of these types of regions were identified. We therefore recommend this index as a better indicator to select priority areas.
Keywords: Conservation index; Ebenopsis caesalpinioides; Ethnobotanical value; Protected Natural Areas; Priority species; Stenocereus martinezii
© 2024 Universidad Nacional Autónoma de México, Instituto de Biología. Este es un artículo Open Access bajo la licencia CC BY-NC-ND
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Áreas prioritarias para la conservación con base en especies de plantas vasculares endémicas y sus atributos bioculturales: un estudio de caso en Sinaloa, México
Resumen
Las plantas vasculares endémicas son uno de los principales indicadores empleados para proponer áreas prioritarias de conservación. La riqueza de especies endémicas y el endemismo ponderado y corregido son frecuentemente incluidos en los análisis, mientras que aspectos antropogénicos o bioculturales como el valor etnobotánico o la vulnerabilidad ecológica son poco considerados. Este trabajo propone áreas prioritarias de conservación para Sinaloa, México, considerando su riqueza de especies endémicas, endemismo ponderado y corregido, así como el valor etnobotánico, estatus de protección e índice prioritario de conservación (IPC). El análisis se realizó en cuadrículas de 19 × 19 km e incluyó 274 registros de 78 especies. Las áreas resultantes, considerando únicamente la riqueza de especies y el endemismo ponderado, coinciden con áreas previamente conocidas por su alta biodiversidad en el estado, mismas que poseen altos esfuerzos de colectas y bajos impactos antropogénicos. Por el contrario, cuando se consideró el valor etnobotánico y el estatus de protección, las áreas prioritarias incluyen zonas con alto impacto antropogénico, pero con presencia de especies con importancia biocultural y valor económico. Empleando el IPC se identificaron ambos tipos de regiones; en consecuencia, recomendamos este índice como un mejor indicador para seleccionar áreas prioritarias.
Palabras clave: Índice de conservación; Ebenopsis caesalpinioides; Valor etnobotánico; Áreas naturales protegidas; Especies prioritarias; Stenocereus martinezii
Introduction
Plants are essential organisms for maintaining the equilibrium of ecosystems and life on Earth. They provide the vast majority of the ecosystem and subsistence services that humans need to survive, including food, medicine, shelter, oxygen, carbon capture, and soil retention. Caring for plants is therefore an act of self-preservation (Raven, 2018). However, over 50% of the terrestrial vegetation on Earth is severely or moderately altered (Bradshaw et al., 2021).
Mexico is the country with the third to fifth highest plant richness, with more than 23,000 species, half of which are endemic (Conabio, 2023a; Villaseñor & Meave, 2022). However, despite 12% of Mexican territory being decreed as Protected Natural Area, it is estimated that between 37 and 50% of the nation’s land area has been impacted by human activities and that the majority of well-conserved areas are located in desert, semi-desert, and high mountain areas that are difficult to access (González-Abraham et al., 2015; Mora, 2019). Two of the largest and most biodiverse ecosystems in the country —dry forest and temperate forest— have suffered total degradation of 37 and 26% of their cover, respectively (Conabio, 2023b; Ulloa-Ulloa et al., 2017). The main causes of this deforestation have been agriculture and infrastructure development, both in Mexico specifically and worldwide (González-Abraham et al., 2015; Laso-Bayas et al., 2022).
One of the main analytical approaches used to propose priority conservation areas is grid analysis, which identifies centers of high biodiversity (“hotspots”) using criteria such as species richness, richness of endemic species, weighted endemism (WE), presence of threatened species, diversity of specific taxa (families or genera), or phylogenetic richness (Gutiérrez-Rodríguez et al., 2022; Maassoumi & Ashouri, 2022; Mehta et al., 2023; Murillo-Pérez et al., 2022; Qin et al., 2022; Sosa & De-Nova, 2012; Vargas-Amado et al., 2020; Villaseñor et al., 2022). The richness of endemic species in particular has the advantage of using a more precise (though smaller) database than the other aforementioned criteria for grid analysis to indicate conservation priority areas.
On the other hand, other indices can be used to propose conservation priority species based on their ethnobotanical or biocultural value, or the degree of threat they face due to use (e.g., Value of Use, Frequency of Use, Conservation Index) (De Lucena et al., 2013; Dhar et al., 2000; Mehta et al., 2023; Pío-León et al., 2023). However, these indices are not usually included in grid richness analyses to select priority conservation areas. These indices weight each species’ value based on its conservation priority, such that a priority conservation area would be determined not just by the total number of species or endemism, but also by their qualities.
Pío-León et al. (2023) compiled a list of the vascular plant species of Sinaloa and proposed some priority conservation areas based on the presence of 2 or more endemic species. In addition, the authors proposed a Priority Conservation Index (PCI) for each species based on its ethnobotanical value and ecological vulnerability, considering characteristics such as its distribution, habitat, and anthropogenic threats. In this index, species with high ethnobotanical value, slow growth (arboreal habit), threatened habitat (near to agricultural zones), and small distribution area (1 or a few known localities), have higher priority than those with no known ethnobotanical value, rapid growth (herbs), inaccessible habitat (cliffs or steep slopes), and wide distribution. The PCI was calculated with the formula:
PCI= D + H + Fv + Am + VE + Vc
where D is distribution; H, habitat; Fv, life form or habit (Spanish abbreviation); Am, degree of threat to their populations; VE, ethnobotanical value, and Vc, commercial value. However, that work did not perform a grid richness analysis to incorporate the values of these indices with traditional algorithms such as WE.
In the present work, we propose priority conservation areas in Sinaloa considering 3 types of algorithms: 1) richness of endemic species, WE, and corrected weighted endemism (CWE); 2) ethnobotanical value, protection status (NOM-059-SEMARNAT-2010 or IUCN) and PCI, and 3) the combination of 1) and 2). We hypothesized that incorporating those anthropogenic and biocultural attributes would modify the priority conservation areas selected since they will not necessarily correspond to the areas of the highest species richness.
Materials and methods
Sinaloa is located in northwestern Mexico, bordered on the east by the Sierra Madre Occidental (SMO) and on the west by the Pacific Ocean. According to Wiken et al. (2011), the main level III ecoregions that compose it are: 1) Sinaloa and Sonora Hills and Canyons with Xeric Shrub and Low Tropical Deciduous Forest (SS-TDF) (50%, 27,568 km2), which is located in the low parts of the SMO; 2) Sinaloa Coastal Plain with Low Tropical Thorn Forest and Wetland (S-TF) (29%, 15,612 km2), located in the lowlands near the coast, from the south-central portion northward; and 3) SMO with Conifer, Oak, and Mixed Forests (PQF) (15.78%, 8,681 km2) in the high parts of the western slope of the SMO (Fig. 1). It has been estimated that 4,000 species of vascular plants, nearly 80 of them endemic, occur in Sinaloa (Pío-León et al., 2023; Vega-Aviña et al., 2021). However, a large part of the coastal territory has been converted to agricultural land (~ 28,000 km2) (INEGI, 2023), resulting in severely fragmented habitats.
The database was based on the file generated by Pío-León et al. (2023) with some updates (Table 1). We incorporated recently described species (until October 2023) and removed species and collections that lacked reliable geographic coordinates. In addition, we prepared a matrix of weighted values considering the ethnobotanical value (E; 1 = documented use, 0 = no documented use), inclusion in a risk category (R) by the NOM-059-SEMARNAT-2010 (Semarnat, 2019) or IUCN (2023) (1 = included in at least 1 category, 0 = not included), and the value of the Conservation Priority Index (PCI), using the values reported by Pío-León et al. (2023) (Table 1). For the PCI, we assigned values according to their quartile position: 4 (upper quartile), 3 (second quartile), 2 (third quartile), and 1 (lower quartile). From these data, we formed 3 analysis groups: 1) biocultural value (E+R; 0 to 2), 2) PCI value (1 to 4), and 3) PCI + R (1 to 5).
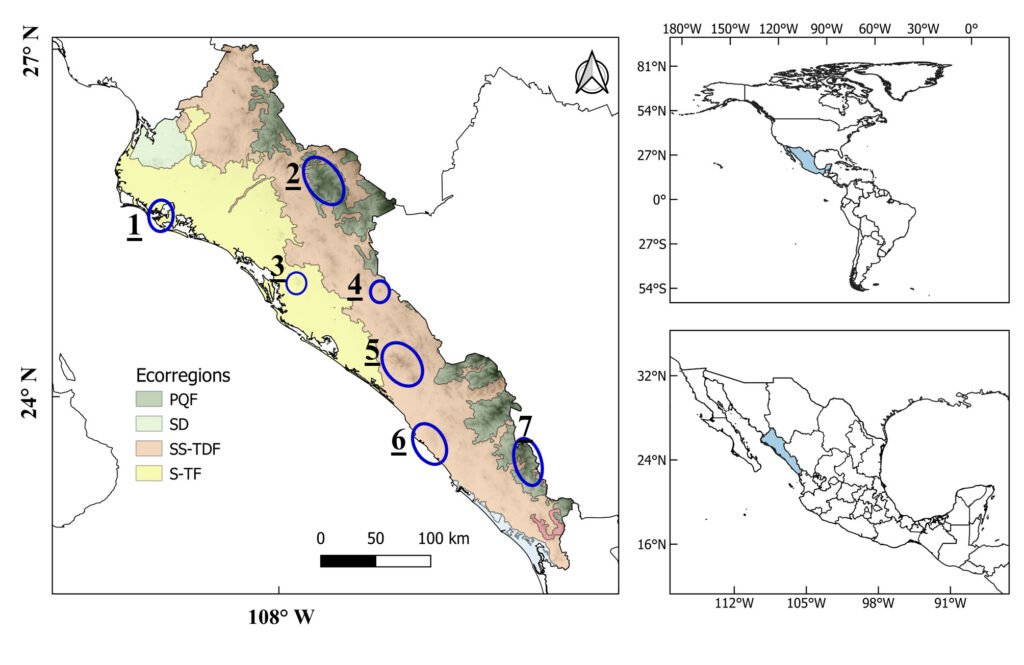
Figure 1. Sinaloa state, Mexico, its main ecoregions level III (Wiken et al., 2011), and the regions of endemism according to Pío-León et al. (2023). Ecoregions: PQF = Conifer, Oak, and Mixed Forests of the Sierra Madre Occidental; SD = Sonoran Desert; SS-TDF = Sinaloa and Sonora Hills and Canyons with Xeric Shrub and Low Tropical Deciduous Forest; S-TF = Sinaloa Coastal Plain with Low Tropical Thorn Forest and Wetlands. Regions of endemism: 1 = Maviri-Topolobampo, 2 = Surutato region, 3 = Cerro Tecomate, 4 = Cerro Colorado, 5 = Sierra Tacuichamona, 6 = Meseta de Cacaxtla, 7 = Sierra de Concordia.
Table 1
List of endemic species of Sinaloa considered for this study and their scores by attributes. E = Ethnobotanical value; R = species with conservation status by the NOM-059-SEMARNAT-2010 or the IUCN (risk); PCI = Priority Conservation Index according to their quartile position.
| Especies | E | R | PCI | E+R | PCI+R |
| Acourtia gentryi L. Cabrera | 0 | 0 | 2 | 0 | 2 |
| Acourtia sinaloana B.L. Turner | 0 | 0 | 1 | 0 | 1 |
| Ageratina concordiana B.L. Turner | 0 | 0 | 2 | 0 | 2 |
| Albizia ortegae Britton & Rose | 0 | 0 | 3 | 0 | 3 |
| Aloysia nahuire A.H. Gentry & Moldenke | 1 | 0 | 4 | 1 | 5 |
| Anemia brandegeei Davenp. | 0 | 0 | 1 | 0 | 1 |
| Arachnothryx sinaloae Borhidi | 0 | 0 | 2 | 0 | 2 |
| Bastardiastrum tarasoides Fryxell | 0 | 0 | 2 | 0 | 2 |
| Bastardiastrum wissaduloides (Baker f.) Bates | 0 | 0 | 1 | 0 | 1 |
| Bletia santosii H. Ávila, J.G. González & Art. Castro | 0 | 0 | 2 | 0 | 2 |
| Bourreria franciscoi Pío-León & Vega | 0 | 0 | 3 | 0 | 3 |
| Bourreria ritovegana Pio-León, M.G. Chávez & L.O. Alvarado | 0 | 0 | 3 | 0 | 3 |
| Bouvardia sinaloae Borhidi & E. Martínez | 0 | 0 | 2 | 0 | 2 |
| Calliandra estebanensis H.M. Hern. | 0 | 0 | 2 | 0 | 2 |
| Carlowrightia fuertensis T.F. Daniel | 0 | 0 | 3 | 0 | 3 |
| Castilleja racemosa (Breedlove & Heckard) T.I. Chuang & Heckard | 0 | 0 | 3 | 0 | 3 |
| Chrysactinia lehtoae D.J. Keil | 0 | 0 | 2 | 0 | 2 |
| Cnidoscolus sinaloensis Breckon ex Fern.Casas | 0 | 1 | 3 | 1 | 4 |
| Cochemiea thomasii García-Mor., Rodr. González, J. García-Jim. & Iamonico | 0 | 0 | 2 | 0 | 2 |
| Coutaportla helgae Pío-León, Torr.-Montúfar & H. Ávila | 0 | 0 | 1 | 0 | 1 |
| Coutaportla lorenceana Torr.-Montúfar, H. Ochot. & Art.Castro | 0 | 0 | 2 | 0 | 2 |
| Croton ortegae Standl. | 0 | 0 | 3 | 0 | 3 |
| Ctenodon rosei Morton | 0 | 0 | 3 | 0 | 3 |
| Cuphea delicatula Brandegee | 0 | 0 | 2 | 0 | 2 |
| Cyclanthera monticola Gentry | 0 | 0 | 2 | 0 | 2 |
| Dioscorea sinaloensis O. Téllez | 0 | 0 | 2 | 0 | 2 |
| Dryopetalon breedlovei (Rollins) Al-Shehbaz | 0 | 0 | 1 | 0 | 1 |
| Ebenopsis caesalpinioides (Standl.) Britton & Rose | 1 | 1 | 4 | 2 | 5 |
| Echeveria coppii Moran ex Gideon F.Sm. & Bischofberger | 0 | 0 | 2 | 0 | 2 |
| Echeveria juliana Reyes, González-Zorzano & Kristen | 0 | 0 | 1 | 0 | 1 |
| Echeveria kimnachii J. Meyrán & R. Vega | 0 | 0 | 1 | 0 | 1 |
| Epidendrum petacaense Hágsater, J. Duarte & Pío-León | 0 | 0 | 2 | 0 | 2 |
| Eryngiophyllum rosei Greenm. | 0 | 0 | 2 | 0 | 2 |
| Frangula surotatensis (Gentry) A. Pool | 0 | 0 | 2 | 0 | 2 |
| Graptopetalum sinaloensis Vega | 0 | 0 | 1 | 0 | 1 |
| Guardiola stenodonta S.F. Blake | 0 | 0 | 1 | 0 | 1 |
| Helicteres vegae Cristóbal | 0 | 0 | 3 | 0 | 3 |
| Heliopsis sinaloensis B.L. Turner | 0 | 0 | 2 | 0 | 2 |
| Table 1. Continued | |||||
| Especies | E | R | PCI | E+R | PCI+R |
| Hofmeisteria sinaloensis Gentry | 0 | 0 | 1 | 0 | 1 |
| Indigofera sinaloensis M. Sousa & Cruz Durán | 0 | 0 | 2 | 0 | 2 |
| Ipomopsis monticola J.M. Porter & L.A. Johnson | 0 | 0 | 2 | 0 | 2 |
| Iresine arenaria Standl. | 0 | 0 | 1 | 0 | 1 |
| Koanophyllum concordianum B.L. Turner | 0 | 0 | 2 | 0 | 2 |
| Lasianthaea gentryi B.L. Turner | 0 | 0 | 2 | 0 | 2 |
| Lasianthaea ritovegana B.L. Turner | 0 | 0 | 1 | 0 | 1 |
| Licania mexicana Lundell | 0 | 0 | 2 | 0 | 2 |
| Lobelia macrocentron (Benth.) T.J. Ayers | 0 | 0 | 2 | 0 | 2 |
| Lopezia conjugens Brandegee | 0 | 0 | 1 | 0 | 1 |
| Lopezia sinaloensis Munz | 0 | 0 | 1 | 0 | 1 |
| Lupinus gentryanus C.P. Sm. | 0 | 0 | 2 | 0 | 2 |
| Lupinus howard-scottii C.P. Sm. | 0 | 0 | 2 | 0 | 2 |
| Lupinus sinaloensis C.P. Sm. | 0 | 0 | 2 | 0 | 2 |
| Mariosousa gentryi Seigler & Ebinger | 0 | 0 | 3 | 0 | 3 |
| Mimosa coelocarpa B.L. Rob. | 0 | 0 | 3 | 0 | 3 |
| Mitracarpus aristatus Borhidi & Lozada-Pérez | 0 | 0 | 2 | 0 | 2 |
| Molinadendron sinaloense (Standl. & Gentry) P.K. Endress | 0 | 1 | 3 | 1 | 4 |
| Pavonia gentryi Fryxell | 0 | 0 | 2 | 0 | 2 |
| Peniocereus papillosus (Britton & Rose) U. Guzmán | 0 | 0 | 2 | 0 | 2 |
| Periptera trichostemon Bullock | 0 | 0 | 2 | 0 | 2 |
| Perityle canescens Everly | 0 | 0 | 1 | 0 | 1 |
| Perityle grandifolia Brandegee | 0 | 0 | 1 | 0 | 1 |
| Perityle stevensii B.L. Turner | 0 | 0 | 1 | 0 | 1 |
| Physalis vestita Waterf. | 0 | 0 | 3 | 0 | 3 |
| Pitcairnia monticola Brandegee | 0 | 0 | 1 | 0 | 1 |
| Polygala polyedra Brandegee | 0 | 0 | 1 | 0 | 1 |
| Psacalium quercifolium H.Rob. & Brettell | 0 | 0 | 2 | 0 | 2 |
| Salvia beltraniorum J.G.González, Pío-León & Art.Castro | 0 | 0 | 2 | 0 | 2 |
| Salvia trichostephana Epling | 0 | 0 | 2 | 0 | 2 |
| Sedum copalense Kimnach | 0 | 0 | 1 | 0 | 1 |
| Stenocereus martinezii (J.G. Ortega) Bravo | 1 | 1 | 4 | 2 | 5 |
| Stevia concordiana B.L. Turner | 0 | 0 | 2 | 0 | 2 |
| Sysyrinchium jacquelineanum Art.Castro, H. Ávila & J.G. González | 0 | 0 | 1 | 0 | 1 |
| Tibouchina thulia Todzia | 0 | 0 | 1 | 0 | 1 |
| Tillandsia mazatlanensis Rauh | 0 | 0 | 2 | 0 | 2 |
| Tillandsia occulta H. Luther | 0 | 0 | 2 | 0 | 2 |
| Verbesina microcarpa S.F. Blake | 0 | 0 | 2 | 0 | 2 |
| Verbesina ortegae S.F. Blake | 0 | 0 | 2 | 0 | 2 |
| Verbesina sinaloensis B.L. Turner | 0 | 0 | 2 | 0 | 2 |
Richness of endemism (SR), weighted endemism (WE), and corrected weighted endemism (CWE). The richness of endemic species was quantified in 19 × 19 km cells (361 km2), dividing Sinaloa into 195 cells. The cell size used was determined according to the criterion of Oyala (2020). Endemic species richness was quantified as the total number of endemic species whose distribution includes the cell. Endemism was evaluated using the WE and CWE indices. The WE score for each cell was obtained by summing, for each species present in the cell, the inverse of the number of cells in which the species occurs; thus, a high WE value indicates cells that contain more species with restricted distributions (i.e., that are found in few other cells), while low WE values indicate cells that mostly contain widely distributed species (i.e., species that are also present in other cells). The CWE is similar, but additionally corrects for potential biases due to differences in overall richness by dividing the value of the WE by the number of species present in the cell (Laffan & Crisp, 2003). The 3 parameters (SR, WE, and CWE) were estimated in the program Biodiverse v.2.0 (Laffan et al., 2010). Geoprocessing of the data was performed in QGIS 3.4.8 (QGIS.org, 2019).
Endemism weighted by biocultural attributes and PCI. In addition to SR, WE, and CWE analysis, endemism weighted by biocultural attributes was evaluated using 2 sets of attribute/parameter combinations, each resulting in 3 maps, 9 in total (Fig. 2). The first set included the species richness plus the biocultural values, resulting in the following 3 combinations: species richness plus biocultural value (SR+E+R), species richness plus PCI (SR+PCI), and PCI plus the risk category (SR+PCI+R). The second set did not consider species richness, resulting in the combinations of biocultural value (E+R), PCI, and PCI+R. For this second set of analyses, only species that fulfilled the relevant criteria were included (e.g., the E+R combination included only species that had ethnobotanical value and are included in a risk category). As such, in the first set of maps, a priority conservation area depended by the number of species present and their qualities (e.g., species with ethnobotanical value or species with protected status), while in the second only the species’ qualities were considered.
The final priority conservation areas were based on the consensus map of the 9 different endemism maps. The consensus areas took into account only the cells that had the highest possible value of the relevant variables in at least 1 of the 9 previously generated endemism maps. The consensus values were obtained by summing the number of times each cell had the highest possible value in each of the endemism maps, such that the highest possible consensus value was theoretically 9 (the cell had the highest possible value in all maps), and the minimum value was 1 (maximal value in only 1 map). The consensus map was also overlayed with Protected Natural Areas and Priority Terrestrial Regions, land use, and bioclimatic corridors.
Results
Occurrence, conservation (risk) status, and ethnobotanical uses of the endemic species of Sinaloa. The database contained 247 records of 78 species, 30 families, and 61 genera. For 48 of the genera (78.7%), only 1 species of the genus was present. The majority of the records were distributed in the central to the southern region of the state, near the coast, in the Meseta de Cacaxtla Natural Protected Area and the area between the former and Sierra de Tacuichamona, as well as in the Concordia and Surutato mountains of the SMO (Fig. 3; regions 6, 5, and 2, in Figure 1). The 2 level III ecoregions best represented were SS-TDF (166 records/ 40 species) and the PQF (53/ 37), followed by the S-TF (17/ 8) (Fig. 3a). Sixty-nine percent of the records fell outside of the polygons of Protected Natural Areas or Priority Terrestrial Conservation Regions (Fig. 3B). Sixty-eight percent of the species (53) were known from a single locality (either a single collection or collections from locations that are very close to each other).
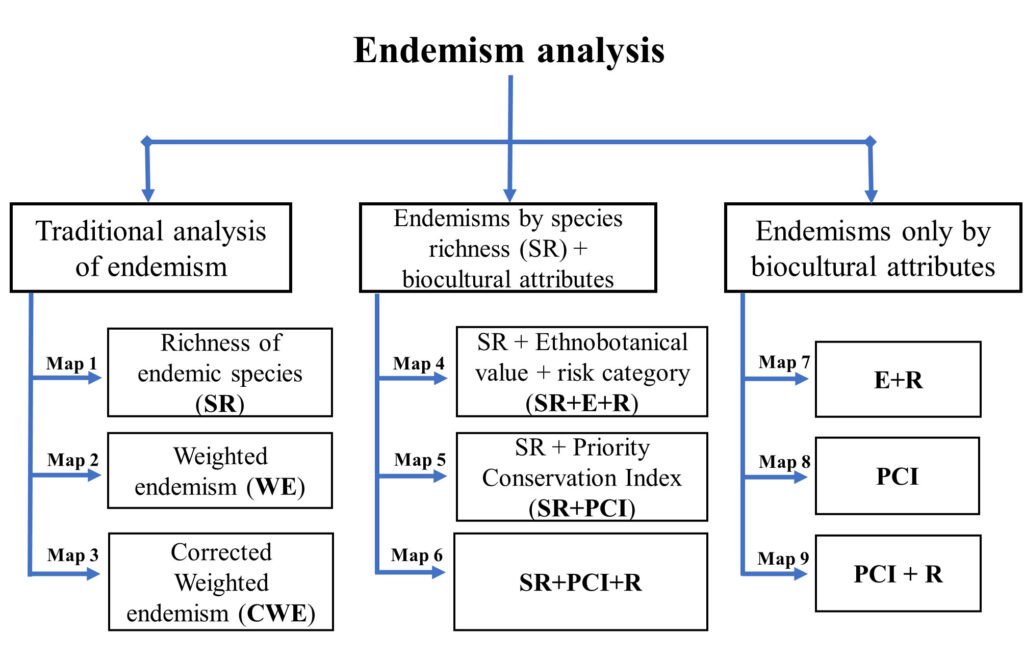
Figure 2. Flowchart of the endemism analysis to select priority areas in Sinaloa, Mexico.
Only 4 species (Cnidoscolus sinaloensis, Ebenopsis caesalpinioides, Molinadendron sinaloense, and Stenocereus martinezii) of the 78 analyzed are found in some risk category (Table 1). All 4 are considered endangered (EN) by the IUCN, while only Stenocereus martinezii is included in NOM-059-SEMARNAT-2010, under the category of special protection (Pr). Only 3 species have well-documented ethnobotanical uses: Aloysia nahuire (aromatic and medicinal tea), Ebenopsis caesalpinioides (edible seeds, occasional commercial value), and Stenocereus martinezii (edible fruits, commercial value). One additional species, Lupinus gentryianus, was noted in the type collection to be used as an anti-parasitic for livestock; however, this plant is only known from that locality, and this use has not since been confirmed, so it was not considered.
The different patterns of endemism are shown in Figure 4. The overall richness of endemism (Fig. 4A) showed 2 main areas —1 in the northern part of the Sierra de Concordia (region 7, Fig. 1) and the other in the western part of the Sierra de Tacuichamona (region 5, Fig. 1)— as well as 3 secondary areas located in the Sierra de Surutato (region 2, Fig. 1), Cerro Colorado (region 4, Fig. 1), and the southern part of the Sierra de Concordia. The WE (Fig. 4B) showed a similar pattern in richness but with an increase in the priority levels of the Sierra de Surutato and a decrease by 1 level for the Tacuichamona and Cerro Colorado. The CWE (Fig. 4C) showed several priority areas more scattered across the state than the WE, mainly in the SMO, corresponding to the majority of the species known from a single locality; however, compared with WE, there was a greater concentration of high-priority cells toward the northern part of the state, near southern Sonora, in the area around the Sierra de Barobampo and Hills of Topolobampo (region 1, Fig. 1).

Figure 3. Records of endemic species in Sinaloa overlayed onto: level III ecoregions (Wiken et al., 2011; definitions in Figure 1) (A) and Protected Natural Areas (PNA) and Priority Terrestrial Regions (B). Categories of Protected Natural Areas: PNAS = state; PNAM = municipal; PNAF = federal; PTR = Priority Terrestrial Regions.
Figure 4. Endemism areas of vascular plants in Sinaloa, Mexico, according to the calculated index values (A-I): SR = endemic species richness; WE = weighted endemism; CWE = corrected weighted endemism; E = ethnobotanical value; R = species with protection status; PCI = Priority Conservation Index.
Figure 5. Consensus priority conservation areas (PAC) in the state of Sinaloa (A-D). Consensus map (A) superimposed to: Protected Natural Areas/Priority Terrestrial Regions (B), land use (C), and bioclimatic corridors (D). Categories of Protected Natural Areas: PNAS = state; PNAM = municipal; PNAF = federal; PTR = Priority Terrestrial Regions.
The addition of the ethnobotanical attributes to the protection status and richness of endemic species (SR+E+R) (Fig. 4D) showed an increase in the values for the areas from the Meseta de Cacaxtla (region 6, Fig. 1) to Sierra de Tacuichamona, but a decrease in the zones of the SMO. Adding the Priority Conservation Index to the richness (SR+PCI) (Fig. 4E) showed an increase and homogenization of the priority in all of the aforementioned regions, while adding protection status (SR+PCI+R) (Fig. 3F) did not significantly modify the areas of importance.
Finally, when considering only the ethnobotanical value plus the protection status (E+R), without considering species richness (i.e., eliminating the species that did not have those attributes), the zone of highest priority was concentrated nearly exclusively in the southern part of the state, within and adjacent to the Meseta de Cacaxtla (Fig. 3G). When considering PCI only or PCI plus risk category, there was again a homogenization of the high priority for the 2 mountainous areas (Surutato and Concordia), Meseta de Cacaxtla, Tacuichamona, and surrounding areas (Fig. 4H, I).
The priority conservation areas, as defined by the consensus among the 9 maps analyzed, were composed of 7 polygons grouped into 3 categories (Fig. 5): 4 cells with a value of 6 (of the maximum possible score of 9) in the northern part of the Sierra de Concordia, northwestern part of Sierra Surutato, Meseta de Cacaxtla, and Sierra de Tacuichamona; 1 with a value of 4 in the area between the Meseta de Cacaxtla and Tacuichamona; and 2 with a value of 3 in the southern part of the Sierra de Concordia and southeastern part of the Sierra de Surutato (Fig. 5A). However, since the 3 areas with a value of 3 or 4 were contiguous with areas with a value of 6, 4 priority conservation areas were proposed: Sierra de Surutato (Fig. 45-a), Sierra de Tacuichamona (Fig. 5A-b), Meseta de Cacaxtla (Fig. 5A-c), and Sierra de Concordia (Fig. 5A-d).
Superimposing the consensus map with the map of existing Protected Natural Areas (Fig. 5B) showed that these 4 consensus areas fall partially within protected areas: 1 federal (Área de Protección de Flora y Fauna Meseta de Cacaxtla, Fig. 5B-c), 1 state (Sierra de Tacuichamona, Fig. 5B-b), and 2 municipal (Reserva Chara Pinta in the Sierra de Concordia and Reserva de Surutato, Fig. 5B-d, B-a, respectively). The Sierra de Concordia also includes part of the terrestrial priority region Río Presidio. Regarding land use, the 2 consensus areas in the SMO were found in mixed pine-oak forest with low impact of agricultural activity (Fig. 5C-a, C-d), while the other 2, located in the Sinaloa and Sonora Hills and Canyons with Xeric Shrub and Low Tropical Deciduous Forest ecoregion, present moderate to high impact from irrigated and rainfed agriculture (Fig. 5C-b, C-c). When considering biological corridors, only the priority area in the Meseta de Cacaxtla overlapped with a bioclimatic corridor.
Discussion
The analyses of richness of endemic species and WE showed higher conservation priority in areas that were previously identified as having high endemism (Pío-León et al., 2023), low anthropogenic impact from agriculture, and which have also historically been subject to concentrated collection efforts (Sierra de Surutato and Sierra de Concordia) (Ávila-González et al., 2019; Gentry 1946; Vega-Aviña et al., 2021). On the other hand, the regions defined based on CWE reflected a high number of species known from a single locality, which could indicate the presence of small islands of endemism in the state or low collection effort. In contrast, the inclusion of the ethnobotanical criteria and protection status (E+R) shows a different pattern from species richness, concentrating high priority scored in an area of transition between the coastal plain of Sinaloa and the hills of Sinaloa and Sonora, near the coast in the center-south of the state. These regions correspond to the transition and ecotone between low tropical deciduous forest and thorn forest, which are strongly impacted by anthropogenic activities (irrigated and rainfed agriculture), suggesting that the species with the highest ethnobotanical importance and with protected status (IUCN or NOM-053-SEMARNAT-2010) are found near human activities that require stronger conservation attention than those located in the high parts of the SMO, where the threats are less severe.
The priority conservation areas indicated by the consensus map (Fig. 5) include the regions with the highest richness of endemic species plus the areas with the highest number of species with biocultural importance. These consensus areas are practically the same as those that were assigned the highest priority values when considering only the Priority Conservation Index (PCI) for each species; as such, this index was the most robust single indicator for selecting priority conservation areas. This index combines ethnobotanical parameters such as species’ uses and economic value with ecological parameters such as their distribution, habit, and habitat. Thus, it covers a broad range of criteria that are useful for defining priority species or areas for conservation.
All the priority conservation areas defined by the consensus map (Fig. 5) except 1 included part of a Protected Area polygon, although only 1 was under federal jurisdiction (Meseta de Cacaxtla). The only cell that did not overlap with a Protected Natural Area was adjacent to the Meseta de Cacaxtla, and it was the cell with the largest area of agriculture. This area is important because it contains the 2 species with the highest ethnobotanical value (Ebenopsis caesalpinioides and Stenocereus martinezii), which are also found in a risk category according to the IUCN and NOM-053-SEMARNAT-2010. This area therefore urgently requires conservation and restoration activities, especially for E. caesalpinioides, whose distribution is limited to the area surrounding this cell (Pío-León et al., 2023). Specifically, we recommend avoiding the conversion from rainfed agriculture to technified irrigated agricultural activities, since these are generally more aggressive toward native vegetation. This area is also important because it is located at the transition between lowland deciduous forest and thorn forest of Sinaloa, which could reflect high endemism, in addition to potentially serving as part of the bioclimatic corridor connecting the 2 most important terrestrial ANPs in the state, Meseta de Cacaxtla (federal) and Sierra Tacuichamona (state).
In the present study, the incorporation of the species’ biocultural parameters modified the priority areas for conservation compared to the areas selected when considering only the richness of endemic species, weighted endemism, or corrected weighted endemism. Specifically, the richness analysis identified priority areas in the mountainous and high-diversity regions of Sinaloa, while the ethnobotanical and ecological factors incorporated zones near the coast that have higher anthropogenic impact. The Conservation Priority Index identified all of these priority regions; for this reason, we propose it as a complete and robust index for identifying priority conservation areas. At the state level, we recommend that conservation and restoration actions be implemented in the area of transition between the low tropical deciduous forest and thorn forest. This area simultaneously presents the highest impact of anthropogenic activities and harbors the most important Sinaloa endemic species in terms of biocultural value and protection status —the “pitaya de Sinaloa” (Stenocereus martinezii) and the “guampinola” or “frutilla” (Ebenopsis caesalpinioides). This area should be considered a priority for both conservation and restoration, which would not have been identified as a priority if only the richness of endemism or CWE had been analyzed.
Acknowledgements
The first author is grateful to the Consejo Nacional de Humanidades, Ciencia y Tecnología (Conahcyt) for the grant awarded as part of the Estancias Posdoctorales por México program (I1200/320/2022). We also thank Jorge David López Pérez for his suggestions on data analysis, and the two anonymous reviewers for their comments and suggestions that improved our manuscript.
References
Ávila-González, H., González-Gallegos, J. G., López-Enríquez, I. L., Ruacho-González, L., Rubio-Cardoza, J., & Castro-Castro, A. (2019). Inventario de las plantas vasculares y tipos de vegetación del Santuario El Palmito, Sinaloa, México. Botanical Sciences, 97, 789–820. https://doi.org/10.17129/botsci.2356
Bradshaw, C. J. A., Ehrlich, P. R., Beattie, A., Ceballos, G., Crist, E., Diamond, J. et al. (2021). Underestimating the challenges of avoiding a ghastly future. Frontiers in Conservation Sciences, 1, 615419. https://doi.org/10.3389/fcosc.2020.615419
Conabio (Comisión Nacional para el Conocimiento y Uso de la Biodiversidad). (2023a). México megadiverso. Comisión Nacional para el Conocimiento y Uso de la Biodiversidad, Ciudad de México. Retrieved 01 December, 2023 from: https://www.biodiversidad.gob.mx/pais/quees
Conabio (Comisión Nacional para el Conocimiento y Uso de la Biodiversidad). (2023b). Ecosistemas de México. Comisión Nacional para el Conocimiento y Uso de la Biodiversidad, Ciudad de México. Retrieved 01 December, 2023 from: https://www.biodiversidad.gob.mx/ecosistemas/ecosismex
Dhar, U., Rawal, R. S., & Upreti, J. (2000). Setting priorities for conservation of medicinal plants ––a case study in the Indian Himalaya. Biological Conservation, 95, 57–65. https://doi.org/10.1016/S0006-3207(00)00010-0
De Lucena, R. F., Lucena, C. M., Araújo, E. L., Alves, Â. G., & Albuquerque, U. P. D. (2013). Conservation priorities of useful plants from different techniques of collection and analysis of ethnobotanical data. Anais da Academia Brasileira de Ciências, 85,169–186. https://doi.org/10.1590/S0001-37652013005000013
Gentry, H. S. (1946). Notes on the vegetation of Sierra Surotato in northern Sinaloa. Bulletin of the Torrey Botanical Club, 73, 451–462. https://doi.org/10.2307/2481592
González-Abraham, C., Ezcurra, E., Garcillán, P. P., Ortega-Rubio, A., Kolb, M., & Bezaury Creel, J. E. (2015). The human footprint in Mexico: physical geography and historical legacies. Plos One, 10, e0121203. https://doi.org/
10.1371/journal.pone.0121203
Gutiérrez-Rodríguez, B. E., Vásquez-Cruz, M., & Sosa, V. (2022). Phylogenetic endemism of the orchids of Megamexico reveals complementary areas for conservation. Plant Diversity, 44, 351–359. https://doi.org/10.1016/j.pld.20
22.03.004
IUCN (International Union for Conservation of Nature). (2023). The IUCN Red List of Threatened Species. Version 2022-2. Retrieved 01 December, 2023 from: https://www.iucnredlist.org
INEGI (Instituto Nacional de Estadística, Geografía e Informática). (2023). Censo agropecuario 2022. Resultados definitivos Sinaloa. Retrieved 25 January, 2024 from: https://www.inegi.org.mx/
Laso-Bayas, J. C., See, L., Georgieva, I., Schepaschenko, D., Danylo, O., Dürauer, M. et al. (2022). Drivers of tropical forest loss between 2008 and 2019. Scientific Data, 9, 1–8. https://doi.org/10.22022/nodes/06-2021.122
Laffan, S. W., & Crisp, M. D (2003) Assessing endemism at multiple spatial scales, with an example from the Australian vascular flora. Journal of Biogeography, 30, 511–520. https://doi.org/10.1046/j.1365-2699.2003.00875.x
Laffan, S.W., Lubarsky, E., & Rosauer, A. F. (2010) Biodiverse, a tool for the spatial analysis of biological and related diversity. Ecography, 33, 643–647. https://doi.org/10.1111/
j.1600-0587.2010.06237.x
Maassoumi, A. A., & Ashouri, P. (2022). The hotspots and conservation gaps of the mega genus Astragalus (Fabaceae) in the Old-World. Biodiversity and Conservation, 31, 2119–2139. https://doi.org/10.1007/s10531-022-02429-2
Mehta, P., Bisht, K., Sekar, K. C., & Tewari, A. (2023). Mapping biodiversity conservation priorities for threatened plants of Indian Himalayan Region. Biodiversity and Conservation, 32, 2263–2299. https://doi.org/10.1007/s10531-023-02604-z
Mora, F. (2019). The use of ecological integrity indicators within the natural capital index framework: The ecological and economic value of the remnant natural capital of México. Journal for Nature Conservation, 47, 72–92. https://doi.org/10.1016/j.jnc.2018.11.007
Murillo-Pérez, G., Rodríguez, A., Sánchez-Carbajal, D., Ruiz-Sánchez, E., Carrillo-Reyes, P., & Munguía-Lino, G. (2022). Spatial distribution of species richness and endemism of Solanum (Solanaceae) in Mexico. Phytotaxa, 558, 147–177. https://doi.org/10.11646/phytotaxa.558.2.1
Oyala, V. (2020). Sistemas de Información Geográfica. Retrie-
ved on December 25th, 2024 from: http://volaya.es/writing
Pío-León, J. F., González-Elizondo, M., Vega-Aviña, R., González-Elizondo, M. S., González-Gallegos, J. G., Salomón-Montijo, B. et al. (2023). Las plantas vasculares endémicas del estado de Sinaloa, México. Botanical Sciences, 101, 243–269. https://doi.org/10.17129/botsci.3076
QGIS Development Team (2019) Geographic Information System, version 3.4.8. QGIS Association. Retrieved 11 December, 2019 from: http://www.qgis.org
Qin, F., Xue, T., Yang, X., Zhang, W., Wu, J., Huang, Y. et al. (2022). Conservation status of threatened land plants in China and priority sites for better conservation targets: distribution patterns and conservation gap analysis. Biodiversity and Conservation, 31, 2063–2082. https://doi.org/10.1007/s10531-022-02414-9
Raven, P. H. (2018). Saving plants, saving ourselves. Plants People Planet, 1, 8–13. https://doi.org/10.1002/ppp3.3
Semarnat (Secretaría de Medio Ambiente y Recursos Naturales). (2019). Norma Oficial Mexicana NOM-059-SEMARNAT-2010. Protección ambiental-Especies nativas de México de flora y fauna silvestres-Categorías de riesgo y especificaciones para su inclusión, exclusión o cambio-Lista de especies en riesgo. Diario Oficial de la Federación, Ciudad de México. Retrieved 01 December, 2023 from: https://www.dof.gob.mx/nota_detalle.php?codigo=5578808&fecha=14/11/2019#gsc.tab=0
Sosa, V., & De-Nova, J. (2012). Endemic angiosperm lineages in Mexico: hotspots for conservation. Acta Botanica Mexicana, 100, 293–315. https://doi.org/10.21829/abm100.2012.38
Ulloa-Ulloa, C., Acevedo-Rodríguez, P., Beck, S., Belgrano, M. J., Bernal, R., Berry, P. E. et al. (2017). An integrated assessment of the vascular plant species of the Americas. Science, 358, 1614–1617. https://doi.org/10.1126/science.aao
0398
Vargas-Amado, G., Castro-Castro, A., Harker, M., Vargas-Amado, M. E., Villaseñor, J. L., Ortiz, E. et al. (2020). Western Mexico is a priority area for the conservation of Cosmos (Coreopsideae, Asteraceae), based on richness and track analysis. Biodiversity and Conservation, 29, 545–569. https://doi.org/10.1007/s10531-019-01898-2
Vega-Aviña, R., Vega-López, I. F., & Delgado-Vargas, F. (2021). Flora nativa y naturalizada de Sinaloa. Culiacán, Sinaloa: Universidad Autónoma de Sinaloa.
Villaseñor, J. L., & Meave, J. A. (2012). Floristics in Mexico today: insights into a better understanding of biodiversity in a megadiverse country. Botanical Sciences, 100, S14–S33. https://doi.org/10.17129/botsci.3050
Villaseñor, J. L., Ortiz, E., & Hernández-Flores, M. M. (2022). The vascular plant species endemic or nearly endemic to Puebla, Mexico. Botanical Sciences, 101, 1207–1221. https://doi.org/10.17129/botsci.3299
Wiken, E., Jiménez-Nava, F., & Griffith, G. (2011). North American Terrestrial Ecoregions-Level III. Montreal, Canada: Commission for Environmental Cooperation. Retrieved Jul 17, 2021 from: http://www3.cec.org/islando
ra/en/item/10415-north-american-terrestrial-ecoregionsle
vel-iii
Mitogenome of the Golden Eagle (Aquila chrysaetos) in northwestern Baja California, Mexico: phylogenetic relationships and genetic variation
Francisco J. García-De León a, Gorgonio Ruiz-Campos b, *, Jesús Roberto Oyervides-Figueroa c, Gonzalo De León-Girón b, Carlos Alberto Flores-López b, Dante Magdaleno-Moncayo d, Alicia Abadía-Cardoso e
a Centro de Investigaciones Biológicas del Noroeste, S.C., Laboratorio de Genética para la Conservación, Av. Instituto Politécnico Nacional 195, Playa Palo de Santa Rita Sur, 23096 La Paz, Baja California Sur, Mexico
b Universidad Autónoma de Baja California, Facultad de Ciencias, Carretera Ensenada-Tijuana Km. 103 s/n, 22860 Ensenada, Baja California, Mexico
c Centro de Investigación Científica y de Educación Superior de Ensenada, Departamento de Acuicultura, Carretera Tijuana -Ensenada 3918, Zona Playitas, 22860 Ensenada, B.C., Mexico
d Universidad Autónoma de Baja California, Facultad de Ingeniería, Arquitectura y Diseño, Carretera Ensenada-Tijuana Km. 103 s/n, 22860 Ensenada, Baja California, Mexico
e Universidad Autónoma de Baja California, Facultad de Ciencias Marinas, Carretera Ensenada-Tijuana Km. 103 s/n, 22860 Ensenada, Baja California, Mexico
*Corresponding author: gruiz@uabc.edu.mx (G. Ruiz-Campos)
Received: 20 August 2024; accepted: 20 February 2025
Abstract
We assembled and annotated the mitochondrial genome of golden eagles from the northwest of Baja California, Mexico, using reference and de novo strategies to analyze the synteny of mitochondrial genes, the phylogenetic relationships, and genetic variation of mitochondrial DNA. The length of the Golden Eagle mitogenome was 17,472 bp (base pairs) with a base composition of A (29.8%), C (32.5%), G (14.0%), and T (23.6%). The mitogenome contains 13 genes coding for the protein complexes coxI II-III, Cytb, cATP 6 and 8, and Nicotinamide Adenine Dinucleotide (NADH 1-6); this arrangement is consistent with the general model of the mitogenome reported in other congeneric members. Mitogenomes of individuals from northwestern Baja California are unique and they differ from the mitogenome of southern California golden eagles in 3 traits: 1) molecule size is 140 bp larger than that previously reported, 2) the addition in the annotation of a region called pseudo control (ψRC), and 3) the annotation in 2 fractions of the coding region for the protein NADH dehydrogenase subunit 3 (ND3). The genetic diversity and phylogenetic analyses of the individual genes and mitogenome support a close genetic relatedness between the golden eagles from northwestern Baja California and southern California region.
Keywords: Mediterranean region; Mitochondrial lineages; Mitochondrial genome; Next generation sequencing; Non-model species
Mitogenoma de águila real (Aquila chrysaetos) en el noroeste de Baja California, México: relaciones filogenéticas y variación genética
Resumen
Ensamblamos y anotamos el genoma mitocondrial del águila real del noroeste de Baja California, México, utilizando estrategias de referencia y de novo para analizar la sintenia de genes mitocondriales, las relaciones filogenéticas y la variación genética del DNA mitocondrial. La longitud del mitogenoma fue 17,472 pb (pares de bases) con una composición de bases de A (29.8%), C (32.5%), G (14.0%) y T (23.6%). El mitogenoma contiene 13 genes codificantes de los complejos proteínicos coxI II-III, Cytb, cATP 6 y 8, y nicotinamida adenina dinucleótido (NADH 1-6); esto fue consistente con el modelo general del mitogenoma reportado en otros congéneres. Los mitogenomas de individuos del noroeste de Baja California son únicos y se diferencian del mitogenoma de individuos del sur de California en 3 rasgos: 1) el tamaño de molécula es 140 pb más grande que el reportado, 2) adición de la región llamada pseudocontrol (ψRC) y 3) anotación en 2 fracciones de la región codificante de la proteína NADH deshidrogenasa subunidad 3 (ND3). La diversidad genética y los análisis filogenéticos de los genes individuales y el mitogenoma respaldan una estrecha relación genética entre las águilas reales del noroeste de Baja California y la región del sur de California.
Palabras clave: Región mediterránea; Linajes mitocondriales; Genoma mitocondrial; Secuenciación de próxima generación; Especies no modelo
Introduction
The Golden Eagle, Aquila chrysaetos, is an accipitrid of boreal distribution that inhabits a variety of open and semi-open biotopes from sea level to 3,630 m in altitude in biomes as tundra, chaparral, temperate grassland, temperate deciduous forest, and coniferous forest (De León-Girón et al., 2016; Flesch et al., 2020; Kochert et al., 2002; Watson et al., 2011). This emblematic species is known to occur in Mexico in the arid and semiarid environments of the northern and central regions, including some locations as south as Oaxaca (Bolger et al., 2014; De León-Girón et al., 2016; Howell & Webb, 1995; Rodríguez-Estrella, 2002; Rodríguez-Estrella et al., 1991, 2020). Although the demography of the Golden Eagle has been determined with different genetic methods that confirm the structuring of its populations (Craig et al., 2016; Doyle et al., 2016), little is known about the genetic identity of populations of this eagle in its distribution range. Northwestern Baja California is considered the greatest nesting and conservation potential region for the Golden Eagle in Mexico (De León-Girón et al., 2016; Rodríguez-Estrella, 2002; Rodríguez-Estrella et al., 1991; Tracey et al., 2017), with vast areas of habitats still pristine for the population conservation of this species shared with the United States of America (Craig et al., 2016; De León-Girón et al., 2016, 2024; Doyle et al., 2014; Katzner et al., 2023).
Studies based on mitochondrial DNA (mtDNA) allow determining the current state of conservation of species, as well as the identification of significant evolutionary and management units with their demographic aspects (Moritz, 1994). With the advent of next-generation sequencing (NGS) technologies (especially on complete mitochondrial genomes), new studies with non-model species became more common. As was the case for various species of the family Accipitridae, including the Golden Eagle. Doyle et al. (2014) were the first to describe the nuclear and mitochondrial genome of the Golden Eagle from the central California region, in the USA.
In this study we focus on the synteny of mitochondrial genes, the phylogenetic relationships, and the mitochondrial genetic diversity among Golden Eagle individuals in northwestern Baja California, Mexico. Given the high migration potential of this species, individuals collected in northwestern Baja California are predicted to be phylogenetically related to the westernmost Golden Eagle population in the USA. Therefore, studies focused on examining the genetic variation at the mitochondrial level will allow better decision-making for management and conservation programs for the species in a binational spectrum. In the same way, our study will be a valuable reference for analyzing other Golden Eagle populations in Mexico and other regions of its distribution range.
Figure 1. Geographical location of the Golden Eagle voucher specimens examined for mitogenome in the northwestern Baja California, Mexico. Geographic coordinates for each individual can be found in Supplementary material T1. Map by Rafael Hernández Guzmán.
Materials and methods
Tissue samples of 6 Golden Eagle specimens from northwestern Baja California, Mexico were obtained. These specimens were found dead in different agriculture valleys of the Mediterranean region in northwestern Baja California, Mexico, between the municipalities of Rosarito and San Quintín, from 1995 to 2014, and deposited as vouchers in the Bird Collection of the Science Faculty, at the Autonomous University of Baja California (UABC), campus Ensenada (Fig. 1, Supplementary material: T1).
For the DNA extraction, tissue samples of the pads of the feet of 5 Golden Eagle specimens referred to here as Ach 1 to Ach 5 were obtained. DNA using the nucleic acid purification method by differential saline precipitation (Aljanabi & Martinez, 1997) was extracted. Both feathers and blood for the Ach 6 sample (Supplementary material: T1) were used and extracted DNA with the Qiagen Blood & Tissue kit. The quality and quantity of the extracted DNA in both cases was evaluated using Nanodrop, Qubit and agarose gel electrophoresis.
Extracted DNA (from 77 to 210 ng/µL per sample quantified in Nanodrop for Ach1-Ach5 individuals and 0.581 ng/µL for Ach6 quantified in Qubit) was purified with SpeedBeads magnetic beads (Thermo-Scientific, Waltham, MA, USA) at a 1.2:1 beads:DNA ratio and resuspended in 50 µl of TLE1X buffer. Libraries for shotgun sequencing were prepared, for which purified DNA from each individual was fragmented by sonication in a Bioruptor® (Diagenode, Liege, Belgium) using 2 rounds, each consisting of 5 cycles of 30 sec of sonication and 30 sec without sonication at the highest setting. DNA fragments were then prepared using the Kapa Biosystems® Hyper Prep Kit (KR0961–v4.15, Roche, Basel, Switzerland), with which end repair and A-tailing were performed, adapters were ligated, and PCR was performed with indexed primers (Glenn et al., 2020) for 14 cycles. Following amplification, fragment size selection was performed by double-dip with SpeedBeads magnetic beads (Thermo-Scientific, Waltham, MA, USA) that allow to preserve fragments between ~250 and ~700 base pairs (bp) for each of the samples, which are the appropriate insert size for the Illumina platform. Sequencing was performed paired end on 2 different platforms. We sequenced the Ach1-Ach5 samples on the Illumina MiSeq V.3 at the Georgia Genomics and Bioinformatics Core (GGBC) to generate 300-bp paired-end fragments, and we sequenced Ach6 on an Illumina HiSeq 4000 at the Oklahoma Medical Research Foundation Clinical Genomics Center to generate 150-bp paired-end fragments.
Bioinformatic analysis. The quality of the raw sequences was evaluated using the FastQC program (Andrews, 2010). Subsequently, the sequences using a standard treatment were filtered using the Trimmomatic software (Bolger et al., 2014) with 4 steps. In the first step, the sequencing adapters and over-represented sequences were removed by means of the ILLUMINACLIP function. The second step discarded sequences with a quality value below 30 QS with the AVGQUAL function. The third step removed fragments below 25 QS with the SLIDINGWINDOW function. The fourth step did an even stricter cleanup using MAXINFO. This step conserved sequences of at least 100 base pairs and was configured to preferentially conserve longer sequences with a value of 0.3. For the Ach 5 specimen, an extended and modified version of the standard treatment was used due to the low quality of sequences. This involved applying the SLIDINGWINDOW function, which modifies the quality value.
Assembly and annotation. Two strategies were followed to assemble the mitochondrial genomes: the reference based and the de novo strategy (Machado et al., 2015, 2018). Mapping against a reference mtDNA genome was performed using the Bowtie2 program version 2.3.4.2 with clean reads (Langmead & Salzberg, 2012). The reference mitochondrial genome was from the same species A. chrysaetos, with the GenBank accession number of KF905228.1. De novo assembly only for samples Ach 2 to Ach 6 was performed using the A5-miseq pipeline (Coil et al., 2015). Due to the large number of reads after the filter (Supplementary material: T2) sample Ach 1 was analyzed with the Velvet software version 1.1 (Zerbino & Birney, 2008). The mitogenome was annotated from the scaffolds obtained from the de novo assemblies. We determined scaffolds longer than 4 Kpb by applying a search with the BLAST tool (Basic Local Alignment Search Tool) of the NCBI (National Center for Biotechnology Information) website. The scaffolds were aligned to achieve greater coverage considering the length of the reference genome used previously. Afterwards, RNAweasel (http://megasun.bch.umontreal.ca/RNAweasel/) was implemented to the identification of tRNA’s (transfer RNA), rRNA’s (ribosomal RNA) and introns and, in turn, MFannot (http://megasun.bch.umontreal.ca/RNAweasel/) for the identification of proteins and open reading frames (Sieber et al., 2018).
In addition to the previous annotation, MITOS web server (Bernt et al., 2013) was used to assist in the annotation of de novo mitochondrial genomes, allowing gene names, tRNA and rRNA secondary structures, and codon usage to be obtained. Finally, a manual curation of the annotations was used to review the 6 existing reading frames with the UGENE software (Okonechnikov et al., 2012). The control region was identified based on 99% similarity with an A. chrysaetos partial control region sequence (EF459579.1). Genome Vx software (Conant and Wolfe, 2008) was used to map the Golden Eagle’s mitogenome, using individual Ach 6 to achieve this analysis.
In the annotation of the de novo assembly, the ND3 protein appeared divided into 2 fractions (see Results), a trait that was not reported by Doyle et al. (2014). Therefore, an experimental verification was required through PCR amplification and its subsequent Sanger sequencing. We designed primers from de novo assembly using the NCBI Primer-BLAST program: ND3Ach-1 (5’GCCTGATACTGGCACTTCGT3’ and 5’CCCTATCAATCTGACCCACCG3’) which generates a 716 bp fragment and ND3Ach-2 (5’CTTCTTCGTCGCTACAGGCT3’ and 5’CCTTCCACCGAACCCACTTAA3’) which generates a 774 bp fragment. Eight Golden Eagle samples from the Ornithological Collection of the Faculty of Sciences of the Autonomous University of Baja California were used for PCR amplification. These samples correspond to the 6 individuals used for sequencing and 2 extra samples that are not part of the mitogenome assembly study. The conditions of the PCR reaction were as follows: 6 min at 94°C, 35 cycles of denaturation, annealing, and extension at 94°C for 30 sec, 51°C for 30 sec and at 72°C for 1.5 min, respectively. A final elongation stage at 70°C for 7 min, and a final conservation stage at 15°C. We sent the amplified fragments for sequencing to the SeqXcel.inc Company in San Diego, California, and analyzed with the ABI PRISM® 3130xl Genetic Analyzer DNA kit.
Synteny and search for polymorphisms. A posteriori synteny analysis was performed between the Golden Eagle mitogenome reported by Doyle et al. (2014) and the newly assembled mitochondrial genome of this study. This analysis was carried out with the MAUVE software (Darling et al., 2004). The same reference mitogenome was used to call SNP’s and INDEL’s for the 6 genomes assembled in this work using the SAMtools program (Li, 2011). Filtering the call quality of the variants involved discarding those with a value less than 20 Qs and keeping those with at least 5X depth (Li, 2011). Each variant was manually evaluated, retaining SNPs and INDELs where at least half of the aligned reads showed the change and verified its existence in at least one other individual (Fridjonsson et al., 2011).
Phylogenetic relationships. Phylogenetic relationships were inferred using the individual gene dataset and the mitogenome. The individual gene fragments analyzed consisted of the ND2, ND3, coxI and Cytb recovered from Aquila chrysaetos individuals from GenBank (Supplementary material: T3) and those produced in this study. The mitogenome analysis for phylogenetic relationships consisted of 10 complete mitogenomes: 6 obtained in this study, 2 reference samples of Aquila chrysaetos (LR822062 from the United Kingdom and NC_024087.1 from California, USA), 1 reference sample classified as Aquila heliaca (NC_035806.1), but has been reported to actually consist of a A. chrsyasetos individual (Sangster & Lukesenburg, 2021) and 1 outgroup (Aquila nipalensis, GenBank accession number NC_045042.1). Both datasets were aligned using the MAFFT 7 algorithm (Katoh & Standley, 2013). Removing the non-conserved regions between the sequences of the multiple alignments was done using the online program Gblocks with the default parameters (Castresana, 2000). The nucleotide substitution model and the mutation rate were determined using JModelTest (Posada, 2008), considering the Bayesian information criterion (BIC).
Two phylogenetic reconstruction methods were used: Maximum Likelihood (ML) and Bayesian analysis. In IQ-TREE (Nguyen et al., 2015) the ML method (Felsenstein, 1981) was run with the DNA substitution model selected by ModelFinder (Kalyaanamoorthy et al., 2017) and 1,000 bootstrap pseudo replicates were performed (Hoang et al., 2017). Bayesian inference was used with MrBayes program (Ronquist et al., 2012); for this, 4 Markov Monte Carlo chains (MCMC) were implemented with a total of 10 million generations, and a sampling every 1,000 generations. To construct the consensus phylogenetic tree, a 25% of burn-in was applied. The support of the nodes was evaluated via the posterior probability values. The results of both approaches were visualized with FigTree 1.4 (Rambaut, 2009). For the mitogenome phylogenetic tree, the dataset on genes that were annotated across the entire mitogenome were partitioned (Supplementary material: T4). To determine the most appropriate DNA substitution model for each gene, the Akaike Information Criterion test implemented in jModelTest2 (Darriba et al., 2012; Guindon & Gascuel, 2003) was used (see selected models for each gene in Supplementary material: T4). The partitioned mitogenome dataset was used to estimate a Bayesian phylogenetic tree in MrBayes (Ronquist et al., 2012). The Bayesian analysis was carried out by running 4 MCMC chains for 5 million generations and saving the trees every 1,000 generations. The consensus phylogenetic tree was constructed using a 10% burn-in.
Genetic diversity. Individual DNA segments from the coxI, Cytb, ND2, and -ND3 genes were used to estimate the genetic diversity parameters for the A. chrysaetos species complex since these were the genes that had a relative amount of A. chrysaetos DNA sequences available in GenBank. Using DNA sequences from the Baja California individuals produced in this study and reference sequences from the species complex (Supplementary material: T3) the following indices were estimated with DnaSP 6 (Rozas et al., 2017): the number of segregating sites (S), number of haplotypes (Nh), haplotype diversity (Hd), and nucleotide diversity (π). Genetic diversity indices were calculated among all sequences included in the respective datasets and within specific groups of sequences according to phylogenetic clades or geographic origin of samples. In addition, the genetic distance (uncorrected p-distance) between and within the phylogenetic clades observed within the mitochondrial genetic lineages in PAUP software was estimated (Swofford, 1993). The clade OTU1 included sequences from a diverse geographical background, including all the DNA sequences of the Baja California golden eagles. In contrast, the A. chrysaetos DNA sequences grouped in the other closely related clade were classified as OTU2. Additionally, the DNA sequence of an individual of A. chrysaetos from California (Southern Sierra Nevada) was analyzed separately to compare its genetic distance with those from Baja California, given the geographical proximity between both populations.
Results
Raw sequencing reads ranged from 3,324,579 (individual Ach 1) to 539,825 (individual Ach 4). The number of total sequences after the filter ranged between 3,251,746 (Ach 1) and 526,875 (Ach4) (Supplementary material: T2A).
Assembly and annotation. There were large differences in assembly between individuals; for example, the average depth in the reference mapping for individual Ach 1 is 30.5X, while for Ach 6 is 168.3X, even though Ach 6 had fewer total reads than Ach 1. Assemblies for individuals Ach 1-5 showed fewer aligned sequences (Supplementary material: T2A). For de novo assembly, the average depth for individual Ach 6 is 8.0X. The longest scaffold generated (17,472 bp) allowed us to assemble the entire mitogenome of the Ach 6 individual, eliminating the need to align multiple scaffolds to recover the full length (Supplementary material: T2B). The data yield was inferior when doing de novo assembly for Ach 1-5 individuals, and none of these individuals allowed the recovery of the complete mitochondrial genome (Supplementary material: T3B).
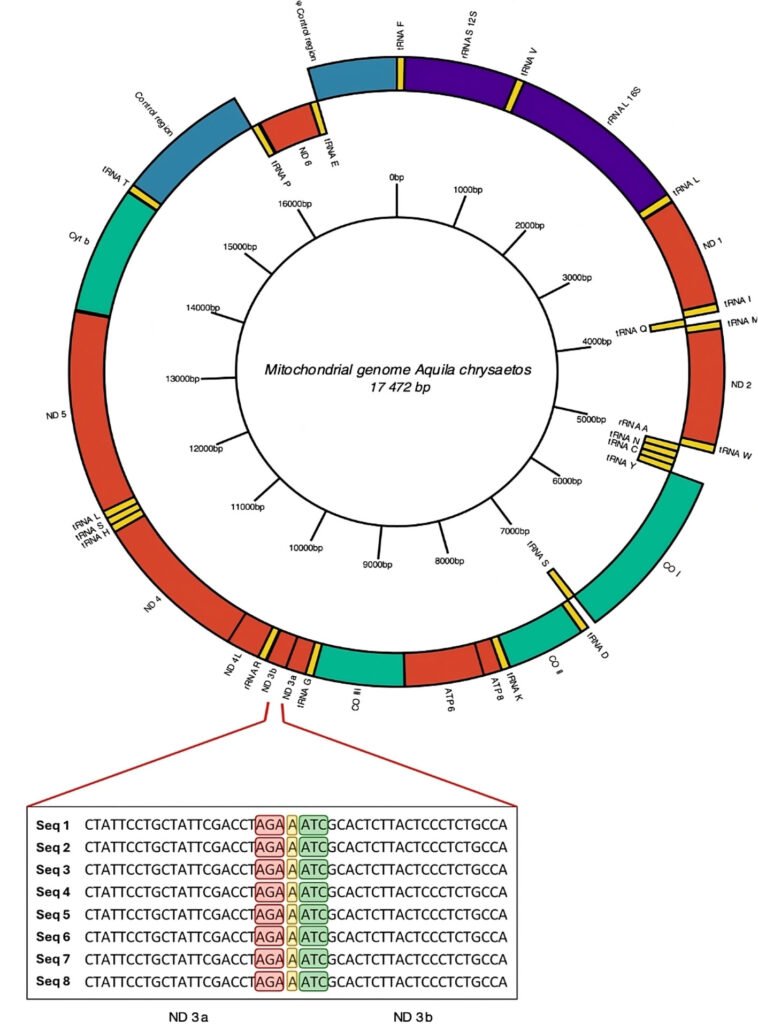
Figure 2. Mitogenome annotated by de novo assembly for Golden Eagle (Ach6) from Baja California, Mexico. In the central part of the diagram in a ring form is shown a scale of the length of mitochondrial genome clockwise. Enlarged view of the section showing the division into 2 fractions of the mitochondrial gene for NADH dehydrogenase subunit 3 (ND3). The stop codon of the ND3a fraction is highlighted in red, the insertion of nucleotide A in yellow, and the start codon of the ND3b fraction in green. The ND3 gene is composed entirely of 352 bp; the ND3a fraction is 207 bp long, while the ND3b fraction is 144 bp long.
Based on the above results, we annotated and curated the mitochondrial genome using only the complete mitogenome obtained by de novo assembly of the Ach 6 individual. All 3 annotation tools identified tRNAs, and none of the tools identified introns within the sequence. The MITOS program (Fig. 2, Supplementary material: T4) was the only software that identified all the genetic elements, such as the 22 tRNAs, 2 rRNAs, and the 13 protein-coding genes that make up the mitogenome. This last annotation was the most complete, so the curation was made from it. We found a region with 7 nucleotide bases that is composed of an AGA stop codon of the ND3 protein, followed by an adenine (A) and finally an ATC start codon that codes for isoleucine. This region divides the ND3 fragment into the 2 regions proposed here (ND3a and ND3b, Fig. 2).
Synteny and search for polymorphisms. The linear order of the mitogenome of Ach 6 coincides with the reference genome (Accession number MT319112.1, Supplementary material: T5). We consider the SNPs and INDELs identified in the Ach 6 reliable, given that the average coverage is 168.3X, and their call quality was high (Supplementary material: T2A). Most SNPs and INDELs are found within genes that code for some protein, transfer RNA and ribosomal genes. The coxI gene presented the greatest number of SNPs with respect to the reference genome in the 6 individuals, while the Cytb and tRNA F genes presented the greatest length changes in the INDELs. In addition, in the ND3 gene, there is a consistent change in the sequences of the 6 individuals with respect to the reference genome (Table 1).
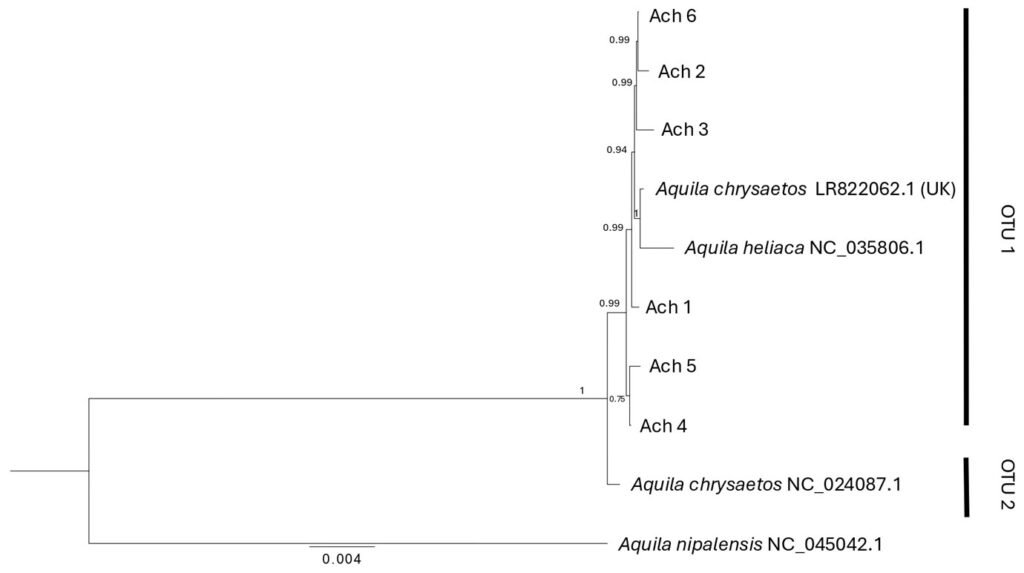
Figure 3. Bayesian phylogenetic tree based on the mitogenome for Golden Eagle voucher specimens from northwestern Baja California, Mexico. Posterior probabilities are shown above internal nodes. GenBank accession numbers are positioned next to reference sequences. Aquila nipalensis was used as an outgroup for rooting the tree. Two Operational Taxonomic Units are identified (OTU 1, 2).
Phylogenetic relationships. With the use of the mitogenome we constructed a phylogenetic tree that identified 2 clades with strong support (1): OTU1 that includes all the individuals analyzed in this study, plus 2 GenBank sequences (Aquila chryseatos, LR82062.1 from the United Kingdom and A. heliatica, NC_035806.1), and OTU2 that corresponds to a GenBank sequence of a specimen collected in California (Fig. 3). For their part, the phylogenetic trees obtained with the fragments of individual genes (ND2, ND3, Cytb and coxI) did not resolve the divergence between individuals from Baja California and California obtained with the mitogenome, but all except the ND3 gene showed a divergence (OTUs 1 and 2), mainly with respect to eagles from the European continent (when information on the collection location is reported, Supplementary material: F1).
Genetic diversity. We observed low genetic diversity, for instance, ND3, ND2, and coxI each had a single segregating site (Table 2), resulting in low nucleotide diversity values (0.0005, 0.0008, and 0.0002, respectively). In contrast, Cytb was the most polymorphic, with a total of 10 segregating sites and a nucleotide diversity an order of magnitude larger (0.00297) (Table 2). The genetic diversity found within OTU1 was higher than that found in OTU2 (Table 2), although OTU1 was not a geographically homogeneous clade among the trees of the different mitochondrial genes, for example in the case of ND2 it was made up of North American eagles, including the individuals of this study, but coxI apart from the previous ones was constituted with eagles from Sweden, Norway and Japan (Table 2).
Discussion
Despite using 2 sequencing platforms for the mitogenome of Golden Eagle from northwestern Baja California, Mexico, we successfully assembled the mitochondrial genome of all 6 individuals by the reference method. This included some individuals (Ach 1-5) with non-consensus regions. Bolger et al. (2014) recommend a pre-processing step of the readings before any analysis, be it assembly by reference or de novo, and mention that if the library and identification adapters are not removed, they can be incorporated in the final assembly. In our case, the pre-processing of the sequences positively influenced the performance of both assemblies and the SNP’s call since the quality of the sequence of the 6 individuals was considerably improved after pre-processing in all aspects of quality reporting.
Table 1
Location of SNP’s and INDEL’s in mitogenomes. Call of SNP’s and INDEL’s for each the Ach 1-6 specimens versus the reference genome of Golden Eagle from the southern Sierra Nevada in California, USA (GenBank accession: KF905228). Pos: Mitogenome position; Ref: nucleotide present in reference sequence. ψCR: Mitochondrial pseudogene control region.
| Pos. | Ref. | Ach1 | Ach2 | Ach3 | Ach4 | Ach5 | Ach6 | Gen |
| SNP’s | ||||||||
| 1,505 | G | A | A | A | ||||
| D-loop | ||||||||
| 1,515 | T | C | ||||||
| 3,130 | G | A | A | A | ||||
| 3,566 | A | G | ND6 | |||||
| 3,695 | T | A | A | A | ||||
| 3,696 | T | A | A | A | ||||
| 5,557 | G | A | A | A | A | A | LSU rRNA | |
| 6,646 | G | A | A | A | ND1 | |||
| 7,141 | A | G | ||||||
| 8,269 | C | T | A | T | ND2 | |||
| 8,593 | A | G | G | A | G | |||
| 8,723 | G | A | A | A | A | tRNA-Trp | ||
| 10,925 | C | T | T | T | T | T | ||
| 10,963 | T | C | ||||||
| 10,970 | C | T | T | T | T | T | ||
| 10,975 | T | C | coxII | |||||
| 11,003 | A | G | G | |||||
| 11,011 | C | T | ||||||
| 11,205 | A | G | ||||||
| 11,213 | C | T | ||||||
| 11,237 | C | T | ||||||
| 11,282 | G | A | ||||||
| 11,284 | G | T | ||||||
| 11,285 | A | C | ||||||
| 11,351 | T | C | ||||||
| 11,354 | A | G | ||||||
| 11,378 | C | T | T | |||||
| 11,432 | C | T | T | T | T | T | ||
| 11,489 | A | C | C | C | C | C | C | |
| 11,493 | T | T | tRNA-Lys | |||||
| 11,496 | G | A | ||||||
| 11,505 | T | G | ||||||
| 17,278 | G | A | A | ND5 | ||||
| INDEL’s | ||||||||
| Table 1. Continued | ||||||||
| Pos. | Ref. | Ach1 | Ach2 | Ach3 | Ach4 | Ach5 | Ach6 | Gen |
| 3 | CTAA | CTAA CTTC CAAA CTAA | Cyt b | |||||
3,550 | T | TGTG AACA A | ψCR | |||||
| 3,701 | CAAA | CCCA CCAA TA | CCAA CAAT AT | tRNA-Phe | ||||
| 13,378 | T | TC | TC | TC | TC | TC | TC | ND3 |
Table 2
Genetic diversity and haplotype composition from gene segments. Tax Set: Group of sequences included in taxonomic set; N: number of sequences; bp: base pairs of DNA sequence included in alignment; S: number of segregating sites; Nh: number of haplotypes; Hd: haplotype diversity; Nd: nucleotide diversity; North America: sequences from either Canada or USA. All sequences from ND3 formed a single clade, and thus were analyzed as a single group. OTUs 1 and 2 in each individual gene refer to the clades detected in each phylogenetic analysis, see Supplementary material F1.
| Mitochondrial sequence | Tax Set | N | bp | S | Nh | Hd | Nd |
| ND3 | All | 11 | 352 | 1 | 2 | 0.182 | 0.00053 |
ND2 | OTU1 | 8 | 1,039 | 0 | 1 | 0 | 0 |
| OTU2 | 4 | 1,039 | 0 | 1 | 0 | 0 | |
| North America | 2 | 1,039 | 0 | 1 | 0 | 0 | |
| Europe | 3 | 1,039 | 0 | 1 | 0 | 0 | |
| All | 12 | 1,039 | 1 | 2 | 0.485 | 0.00087 | |
coxI | OTU1 | 13 | 1,551 | 1 | 2 | 0.154 | 0.00025 |
| OTU2 | 2 | 1,551 | 0 | 1 | 0 | 0 | |
| North America | 4 | 1,551 | 0 | 1 | 0 | 0 | |
| Europe | 4 | 1,551 | 1 | 2 | 0.5 | 0.00082 | |
| All | 15 | 1,551 | 1 | 2 | 0.133 | 0.00022 | |
Cytb | OTU1 | 13 | 1,143 | 1 | 2 | 0.154 | 0.00016 |
| OTU2 | 3 | 1,143 | 1 | 2 | 0.667 | 0.00069 | |
| North America | 4 | 1,143 | 1 | 2 | 0.5 | 0.00052 | |
| Europe | 5 | 1,143 | 9 | 3 | 0.02688 | 0.00541 | |
| All | 16 | 1,143 | 10 | 4 | 0.442 | 0.00297 |
The most reliable mitogenome assembly was that of Ach 6 individual due to its higher alignment rate, its greater depth, and its 100% coverage of the reference genome. This ensemble can be considered true and not an artifact due to the procedures used, such as sampling every 500 generations and a burn-in value of 600,000 generations (Lerner & Mindell, 2005). In addition, we analyzed the data with Bowtie2, which implements an alignment strategy based on the FM-Index and Burrows-Wheeler. These programs have been shown to work well for applications such as INDEL discovery (Lindner & Friedel, 2012) and for aligning long sections of the reference genome (Thankaswamy-Kosalai et al., 2017). Hunt et al. (2014) pointed out that the best result of assembling a genome de novo is when it is contained in a single scaffold for the entire mitochondrial DNA molecule or for each chromosome in the case of the nuclear genome. The Golden Eagle mitochondrial genome assembly of individual Ach 6 was the best because it was found within a single scaffold (17,472 bp) and its depth was at least 8.0X (Supplementary material: T2B; Baker, 2012).
The extension and position of the genes in the mitogenome of the Golden Eagle described in this study (Fig. 2, Supplementary material: T4) showed differences compared to those reported by Doyle et al. (2014) from an individual sampled in central California. For example, the size of the mitogenome described here is larger (17,472 bp) than the one reported by Doyle et al. (2014) (17,332 bp), with a difference of 140 bp. When experimentally analyzing the size of the ND3 gene amplified fragments, the size was between 700 and 800 bp, that is, the theoretically expected size. Another difference consisted in the fraction of the gene encoding the ND3 protein reported here as split into 2 segments (ND3a and ND3b). The distinction of the 2 subunits is, within the reading frame, a region made up of 7 nucleotides between the ATC start codon that starts the protein, and the TAA stop codon that marks the end of the protein. The first 3 of these 7, code for a new AGA stop codon, followed by an adenine (A) nucleotide and 3 nucleotides that code for a new ATC start codon. This causes 2 coding portions to coexist, annotated as ND3, separated by a single nucleotide that changes the reading frame, causing the second portion to appear with its start and stop codons. Mindell et al. (1998) report a nucleotide that is not translated within the ND3 protein sequence. Eberhard and Wright (2016) mentioned that such a trait is observed in the entire order Psittaciformes (parrots, parakeets, and allies).
Specifically, a variety of distinct indels have been found within several of the mitochondrial protein-coding genes in Psittaciformes (Eberhard & Wright, 2016), with variations in terms of their evolutionary origin since some are present in all Psittaciformes and some are more recent in origin and only found within specific taxa. Slack et al. (2003) reported variation in the length of the mitochondrial ND6 gene in other avian taxa. Overall, the biological implications of these variations remain unclear until more studies on the proteins are performed to determine if the protein compositions become altered by these variations. However, the experimental verification carried out in the present study strengthens our annotation as 2 fractions for the ND3 region of the A. chrysaetos mitogenome. Also, we noted the regulatory non-coding region (the pseudo control region, ψCR) to be highly conserved among the A. chrysaetos individuals analyzed, which is a relatively conserved region within the order Accipitriformes (cf. Song et al., 2015) and is also found in the same position between the tRNA -E and tRNA-F, as shown by Liu et al. (2017) for the mitogenome of Accipiter gularis, another congeneric species.
Synteny and search for polymorphisms. The assembled Golden Eagle mitogenome did not present changes in gene order with respect to the reference (KF905228.1, a male Golden Eagle from southern Sierra Nevada, California). As expected, neither did it present changes in the order of the genes with respect to other species of the same group of birds (Accipitridae), such as Buteo buteo (Haring et al., 2001), Accipiter virgatus (Song et al., 2015), Aquila fasciata (Jiang et al., 2015) and Accipiter gularis (Liu et al., 2017). In addition, it is consistent with one of the general models of the birds’ mitogenome that is characterized by a duplication of the control region and other adjacent genes, which were subsequently degraded, giving rise to the ψCR (Eberhard & Wright, 2016). SNP’s and INDEL’s observed in the mitogenome of the Ach 6 individual meet the needed quality (probability of not being an error) and sequence depth (Li, 2011), so they can be considered as potential markers that should be tested in future population studies, considering some technical aspects, and thus avoid the call of false positives (The 1000 Genomes Project Consortium, 2015).
Reconstruction of phylogenetic relationships. The phylogenetic tree classified all Golden Eagle individuals into 2 mitogenome clades (OTU1 and OTU2). All individuals from Baja California, an individual from the United Kingdom, and an A. heliaca individual clustered within OTU1 (Fig. 3), while OTU2 was composed of a single Golden Eagle individual from California. The genetic distance between the 2 OTUs was 0.002 nucleotide differences per site (Supplementary material: T6). This pattern is not surprising given the poor sampling of taxa for both the mitogenomes and individual genes of A. chrysaetos. Once additional mitogenomes are available for this species, it will become possible to increase the phylogenetic resolution that could confirm or reject the potential presence of multiple mitochondrial lineages circulating within Golden Eagle populations. Nevertheless, given the broad geographic area included in the mitogenome dataset (i.e., an A. chrysaetos individual from the United Kingdom, as well an A. heliaca individual), and the low genetic diversity observed within these mitochondrial sequences (see below), suggests that at the mitochondrial level, the genetic diversity found within A. chrysaetos populations is low.
The use of genes or fragments of the mitochondrial genome allows for the analysis to include a larger data set in the number of locations and individuals. The topology of 3 of the 4 individual gene trees (ND2, Cytb and coxI) did support the presence of at least 2 mitochondrial clades (Supplementary material: F1). However, these gene trees were not consistent when grouping the North American Golden Eagle individuals on the same OTUs.
Future studies targeting molecular markers with a higher mutational rate (i.e., microsatellites) or genomics approach could potentially uncover the presence of genetic differences that are not well defined at the mitochondrial level but might be biologically important. One such difference is seen in the species’ bivalent behavior and its migration pattern in the west (Mcintyre & Lewis, 2016) and northwest of the USA, and through Canada (Bedrosian et al., 2018). The mitogenomic lineages that potentially infer the difference between the eagles of Baja California and central California could correspond to the area where 2 ecological populations are coexisting (De León-Girón et al., 2016). The first population is migratory, originating in western North America (Oregon), while the second lineage is resident in Baja California. According to Craig et al. (2016), this lineage has 1 of the less frequent haplotypes of the mitochondrial control region and is restricted to California. The ability of the Golden Eagle to adapt to different environments (Judkins & Van Den Bussche, 2018), the large dispersal distances of reproductive or floating between Mexico and the USA (De León-Girón et al., 2016, 2024; Rodríguez-Estrella et al., 2020; Tracey et al., 2017), and its extensive home range of reproductive pairs (D’Addario et al., 2019), would support these hypotheses. Besides, the presence of these groups of individuals (reproductive and non-reproductive) in Baja California would be part of the reproductive behavior of the species (Watson et al., 2011), that is, the process of succession and substitution of reproductive pairs in the region (De León-Girón et al., 2016).
Genetic diversity. The values of mitochondrial genetic diversity calculated for most genes (except Cytb) were near zero (Table 2). We expected this result, considering that they are genes that code for proteins with highly conserved regions (Dawnay et al., 2007) and are subject to biochemical limitations that cause high levels of homoplasy (Faria et al., 2007). However, Bates et al. (2003) mentioned that ND2 is genetically more diverse than Cytb, while Faria et al. (2007) stated that ND2 is one of the most variable mitochondrial genes within birds and, therefore, regularly implemented in population genetics. A pattern we did not observe in A. chrysaetos since Cytb was the gene with the highest genetic diversity observed, with a haplotype diversity of 0.154 and 0.667 for OTU1 and OTU2, respectively, while both OTUs had a haplotype diversity of zero for the ND2 gene (Table 2).
Implications for conservation. Golden eagles are recognized for presenting large expanses of territory, and their resident, migratory and floating populations (reproductive adults without territories), promote the population gene flow (Craig et al., 2016; Poessel et al., 2022). The mitochondrial DNA data produced in this study confirm a very close genetic relationship between the northwestern Baja California individuals and those from the USA. Therefore, the execution of bi-national conservation programs by the agencies of both countries (SEMARNAT and USFWS) are priorities for the “Californian-Baja Californian” metapopulation of Golden Eagle. It is necessary to increase the number of young individuals with satellite tracking in the southern Baja California peninsula, to continue the genetic characterization (with different types of markers, mitochondrial DNA, microsatellites and SNPs obtained by Next Generation Sequencing) and population monitoring to evaluate the structure and connectivity of the species in both countries.
Our findings warrant developing joint conservation efforts between the governments of Mexico and the USA to monitor and preserve the North American Golden Eagle.
Acknowledgements
Funding for the sequencing was covered by the SAGARPA-INAPESCA PIDETEC project 2017/0647. The authors thank Travis Glen and Natalia Juliana Bayona Vásquez for their Illumina sequencing services. We thank 3 anonymous reviewers who helped improve the manuscript.
References
Aljanabi, S. M., & Martinez, I. (1997). Universal and rapid salt-extraction of high-quality genomic DNA for PCR-based techniques. Nucleic Acids Research, 25, 4692–4693. https://doi.org/10.1093/nar/25.22.4692
Auton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., Korbel, J. O. et al. (2015). A global reference for human genetic variation. The 1000 Genomes Project Consortium. Nature, 526, 68–74. https://doi.org/10.1038/nature15393
Baker, M. (2012). De novo genome assembly: What every biologist should know. Nature Methods, 9, 333–337. https://doi.org/10.1038/nmeth.1935
Bates, J. M., Tello, J. G., & Da Silva, J. M. C. (2003). Initial assessment of genetic diversity in ten bird species of South American Cerrado. Studies on Neotropical Fauna and Environment, 38, 87–94. https://doi.org/10.1076/snfe.38.2.87.15924
Bernt, M., Donath, A., Jühling, F., Externbrink, C., Florentz, G., & Fritzsch, J. (2013). MITOS: Improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution, 69, 313–319. https://doi.org/10.1016/j.ympev.2012.08.023
Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in Phylogenetic Analysis. Molecular Biology and Evolution, 17, 540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Coil, D., Jospin, G., & Darling, A. E. (2015). A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics, 31, 587–589. https://doi.org/10.1093/bioinformatics/btu661
Conant, G. C., & Wolfe, K. H. (2008). GenomeVx: Simple web based creation of editable circular chromosome maps. Bioinformatics, 24, 861–862. https://doi.org/10.1093/bioinformatics/btm598
Craig, E. H., Adams, J. R., Waits, L. P., Fuller, M. R., & Whittington, D. M. (2016). Nuclear and mitochondrial DNA analyses of golden eagles (Aquila chrysaetos canadensis) from three areas in western North America; initial results and conservation implications. Plos One, 11, e0164248. https://doi.org/10.1371/journal.pone.0164248
D’Addario, M., Monroy-Vilchis, O., Zarco-González, M. M., & Santos-Fita, D. (2019). Potential distribution of Aquila chrysaetos in Mexico: Implications for conservation. Avian Biology Research, 12, 33–41. https://doi.org/10.1177/1758155918823424
Darling, A. C. E., Mau, B., Blattner, F. R., & Perna, N. T. (2004). Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Research, 14, 1394–1403. https://doi.org/10.1101/gr.2289704
Darriba, D., Taboada, G., Doallo, R., & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772. https://doi.org/10.1038/nmeth.2109
Dawnay, N., Ogden, R., McEwing, R., Carvalho, G. R., & Thorpe, R. S. (2007). Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Science International, 173, 1–6. https://doi.org/10.1016/j.forsciint.2006.09.013
De León-Girón, G., Rodríguez-Estrella, R., & Ruiz-Campos, G. (2016). Estatus de distribución actual del águila real (Aquila chrysaetos) en el noroeste de Baja California, México. Revista Mexicana de Biodiversidad, 87, 1328–1335. https://doi.org/10.1016/j.rmb.2016.10.003
De León-Girón, G., Toscano, D., Ruiz-Campos, G., Porras-Peña, C., Escoto-Rodríguez, F., Sánchez-Sotomayor, V. G. et al. (2024). Aerial identification and quantification of nesting sites of Golden Eagle (Aquila chrysaetos) in the mountain ranges of central Baja California Peninsula, Mexico. Revista Mexicana de Biodiversidad, 95, e955247.https://doi.org/10.22201/ib.20078706e.2024.95.5247
Doyle, J. M., Katzner, T. E., Bloom, P. H., Ji, Y., Wijayawardena, B. K., & DeWoody, J. A. (2014). The genome sequence of a widespread apex predator, the golden eagle (Aquila chrysaetos). Plos One, 9, e95599. https://doi.org/10.1371/journal.pone.0095599
Doyle, J. M., Katzner, T. E., Roemer, G. W., Cain, J. W., Millsap, B. A., McIntyre, C. L. et al. (2016). Genetic structure and viability selection in the golden eagle (Aquila chrysaetos), a vagile raptor with a Holarctic distribution. Conservation Genetics, 17, 1307–1322. https://doi.org/10.1007/s10592-016-0863-0
Eberhard, J. R., & Wright, T. F. (2016). Rearrangement and evolution of mitochondrial genomes in parrots. Molecular Phylogenetics and Evolution, 94, 34–46. https://doi.org/10.1016/j.ympev.2015.08.011
Faria, P. J., Baus, E., Morgante, J. S., & Bruford, M. W. (2007). Challenges and prospects of population genetic studies in terns (Charadriiformes, Aves). Genetics and Molecular Biology, 30, 681–689. www.sbg.org.br
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution, 17, 368–376. https://doi.org/10.1007/BF01734359
Flesch, A. D., Rodríguez-Estrella, R., Gallo-Reynoso, J. P., Armenta-Méndez, L., & Montiel-Herrera, M. (2020). Distribution and habitat of the Golden Eagle (Aquila chrysaetos) in Sonora, Mexico. Revista Mexicana de Biodiversidad, 91, 1892–2019. https://doi.org/10.22201/ib.20078706e.2020.91.3056
Fridjonsson, O., Olafsson, K., Tompsett, S., Bjornsdottir, S., Consuegra, S., Knox, D. et al. (2011). Detection and mapping of mtDNA SNPs in Atlantic salmon using high throughput DNA sequencing. BMC Genomics, 12, 179. https://doi.org/10.1186/1471-2164-12-179.
Glenn, T. C., Nilsen, R. A., Kieran, T. J., Sanders, J. G., Bayona-Vásquez, N. J., Finger, J. W. et al. (2020). Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). PeerJ, 7, e7755. https://doi.org/10.7717/peerj.7755
Guindon, S., & Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52, 696–704. https://doi.org/10.1080/10635150390235520
Haring, E., Kruckenhauser, L., Gamauf, A., Riesing, M. J., & Pinsker W. (2001). the complete sequence of the mitochondrial genome of Buteo buteo (Aves, Accipitridae) indicates an early split in the phylogeny of raptors. Molecular Biology and Evolution, 18, 1892–1904. https://doi.org/10.1093/oxfordjournals.molbev.a003730
Hoang, D. T., Chernomor, O., Von Haeseler, A., Quang Minh, B., Sy Vinh, L., & Rosenberg, M. S. (2017). UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution, 35, 518–522. https://doi.org/10.5281/zenodo.854445
Howell, S., & Webb, S. (1995). A guide of the birds of Mexico and northern Central America. Oxford, UK: Oxford University Press.
Hunt, M., Newbold, C., Berriman, M., & Otto, T. D. (2014). A comprehensive evaluation of assembly scaffolding tools. Genome Biology, 15, R42. https://doi.org/10.1186/gb-2014-15-3-r42
Jiang, L., Chen, J., Wang, P., Ren, Q., Yuan, J., Qian, C. et al. (2015). The mitochondrial genomes of Aquila fasciata and Buteo lagopus (Aves, Accipitriformes): sequence, structure and phylogenetic analyses. Plos One, 10, e0141037. https://doi.org/10.1371/journal.pone.0136297
Judkins, M. E., & Van Den Bussche, R. A. (2018). Holarctic phylogeography of golden eagles (Aquila chrysaetos) and evaluation of alternative North American management approaches. In Biological Journal of the Linnean Society, 123, 471–482. https://doi.org/10.1093/biolinnean/blx138
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., & Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587–589. https://doi.org/10.1038/nmeth.4285
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software Version 7: improvements in performance and usability article fast track. Molecular Biology and Evolution, 30, 772–780. https://doi.org/10.1093/molbev/mst010
Katzner, T., Kochert, M., Steenhof, K., McIntyre, C., Craig, E., & Miller, T. (2023). Golden Eagle (Aquila chrysaetos), version 2.0. InP. G. Rodewald, & B. K. Keeney (Eds.), Birds of the World. Ithaca, NY: Cornell Lab of Ornithology, USA. https://doi.org/10.2173/bow.goleag.02
Kochert, M. N., Steenhof, K., Mcintyre, C. L., & Craig, E. H. (2002). Golden Eagle (Aquila chrysaetos). In A. Poole, & F. Gill (Eds.), The birds of North America, No. 684 (pp. 1–44). Philadelphia, PA: The Birds of North America, Inc.
Langmead, B., & Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9, 357–359. https://doi.org/10.1038/nmeth.1923
Lerner, H. R. L., & Mindell, D. P. (2005). Phylogeny of eagles, Old World vultures, and other Accipitridae based on nuclear and mitochondrial DNA. Molecular Phylogenetics and Evolution, 37, 327–346. https://doi.org/10.1016/j.ympev.2005.04.010
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. https://doi.org/10.1093/bioinformatics/btr509
Lindner, R., & Friedel, C. C. (2012). A comprehensive evaluation of alignment algorithms in the context of RNA-Seq. Plos One, 7,e52403. https://doi.org/10.1371/journal.pone.0052403
Liu, G., Li, C., Du, Y., & Liu X. (2017). The complete mitochondrial genome of Japanese sparrowhawk (Accipiter gularis) and the phylogenetic relationships among some predatory birds. Biochemical Systematics and Ecology, 70,116–125. https://doi.org/10.1016/j.bse.2016.11.007
Machado, D. J., Janies, D., Brouwer, C., & Grant T. (2018). A new strategy to infer circularity applied to four new complete frog mitogenomes. Ecology and Evolution, 8, 4011–4018. https://doi.org/10.1002/ece3.3918
Machado, D. J., Lyra, M. L., & Grant, T. (2016). Mitogenome assembly from genomic multiplex libraries: comparison of strategies and novel mitogenomes for five species of frogs. Molecular Ecology Resources, 16, 686–693. https://doi.org/10.1111/1755-0998.12492
McIntyre, C. L., & Lewis, S. B. (2016). Observations of migrating golden eagles (Aquila chrysaetos) in eastern interior Alaska offer insights on population size and migration monitoring. The Journal of Raptor Research, 50, 254–264. https://doi.org/10.3356/jrr-15-13.1
Mindell, D. P., Sorenson, M. D., & Dimcheff, D. E. (1998). An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Molecular Biology and Evolution, 15, 1568–1571. https://doi.org/10.1093/oxfordjournals.molbev.a025884
Moritz, C. (1994). Applications of mitochondrial DNA analysis in conservation: a critical review. Molecular Ecology, 3,
401–411. https://doi.org/10.1111/j.1365-294X.1994.tb00080.x
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., & Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. https://doi.org/10.1093/molbev/msu300
Okonechnikov, K., Golosova, O., Fursov, M., Varlamov, A., Vaskin, Y., Efremov, I. et al. (2012). Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics, 28, 1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Poessel, S. A., Woodbridge, B., Smith, B. W., Murphy, R. K., Bedrosian, B. E., Bell, D. A. et al. (2022). Interpreting long-distance movements of non-migratory golden eagles: Prospecting and nomadism? Ecosphere, 13,e4072. https://doi.org/10.1002/ecs2.4072
Posada, D. (2008). jModelTest: phylogenetic model averaging. Molecular Biology and Evolution, 25,1253–1256. https://doi.org/10.1093/molbev/msn083
Rambaut, A. (2010). FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/
Rodríguez-Estrella, R. (2002). A survey of golden eagles in Northern Mexico in 1984 and recent records in Central and Southern Baja California Peninsula. Journal of Raptor Research, 36, 3–9. https://digitalcommons.usf.edu/jrr/vol36/iss5/3
Rodríguez-Estrella, R., De León-Girón, G., Nocedal, J., Chapa, L., Scott, L., Eccardi, F. et al. (2020). Informe del Programa de monitoreo del águila real en México. Ciudad de México: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad/Centro de Investigaciones Biológicas del Noroeste (CIBNOR).
Rodríguez-Estrella, R., Llinas-Gutiérrez, J., & Cancino, J. (1991). New Golden Eagle records from Baja California. Journal Raptor Research, 25, 68–71. https://digitalcommons.usf.edu/jrr/vol25/iss3/3
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S. et al. (2012). Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. https://doi.org/10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sánchez-Del Barrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E. et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34, 3299–3302. https://doi.org/10.1093/molbev/msx248
Sangster, G., & Luksenburg, J. A. (2021). Sharp increase of problematic mitogenomes of birds: causes, consequences, and remedies. Genome Biology and Evolution, 13, 1–14. https://doi.org/10.1093/gbe/evab210
Sieber, P., Platzer, M., & Schuster, S. (2018). The definition of open reading frame revisited. Trends in Genetics, 34, 167–170. https://doi.org/10.1016/j.tig.2017.12.015
Slack, K. E., Janke, A., Penny, D., & Arnason, U. (2003). Two new avian mitochondrial genomes (penguin and goose) and a summary of bird and reptile mitogenomic features. Gene, 302, 43–52. https://doi.org/10.1016/S0378111902010533
Song, X., Huang, J., Yan, C., Xu, G., Zhang, X., & Yue, B. (2015). The complete mitochondrial genome of Accipiter virgatus and evolutionary history of the pseudo-control regions in Falconiformes. Biochemical Systematics and Ecology, 58, 75–84. https://doi.org/10.1016/j.bse.2014.10.013
Thankaswamy-Kosalai, S., Sen, P., & Nookaew, I. (2017). Evaluation and assessment of read-mapping by multiple next-generation sequencing aligners based on genome-wide characteristics. Genomics, 109, 186–191. https://doi.org/10.1016/j.ygeno.2017.03.001
Tracey, J. A., Madden, M. C., Sebes, J. B., Bloom, P. H., Katzner, T. E., & Fisher, R. N. (2017). Biotelemetery data for Golden Eagles (Aquila chrysaetos) captured in coastal southern California, February 2016-February 2017. U.S. Geological Survey Data Series 1051. https://doi.org/10.3133/ds1051
Watson, J., Riley, H., & Thomson, D. (2011). The Golden Eagle (2nd Ed.). New Haven, Connecticut: Yale University Press.
Zerbino, D. R., & Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research, 18, 821–829. https://doi.org/10.1101/gr.074492.107

Avifauna of cloud forest riparian corridors in a degraded landscape in eastern Mexico
Omar A. Hernández-Dávila a, *, Vinicio Sosa b, Javier Laborde b
a Instituto de Biotecnología y Ecología Aplicada, Av. de las Culturas Veracruzanas No. 101, Col. Emiliano Zapata, 91090 Xalapa, Veracruz, Mexico
b Instituto de Ecología, A.C., Red de Ecología Funcional, Carretera Antigua a Coatepec, No. 351, El Haya, 91073 Xalapa, Veracruz, Mexico
*Corresponding author: omar.hernandez.davila@outlook.com (O.A. Hernández-Dávila)
Abstract
Cloud forests are known for their remarkable biodiversity and provide many ecosystem services. However, this biodiversity is in jeopardy due to the conversion of forests to other land uses. At its northernmost range in the Neotropics, cloud forest persists in remnant fragments immersed in an agricultural matrix that still has arboreal elements, such as riparian corridors. In this study we characterize the avifauna present in cloud forest riparian corridors in a highly degraded landscape of Mexico. We classified the avifauna in terms of migratory and conservation status, trophic guild, body mass, forest stratum and habitat preference. In 14 riparian corridors we recorded 86 bird species (75% were resident). Insectivorous and frugivorous species represented 79% of total richness. Almost 65% of species prefer the mid-story or canopy forest strata, while 46% were habitat generalists. Despite crossing open agricultural areas, cloud forest riparian corridors still harbor a diverse assemblage of bird species, which includes not only those tolerant to disturbance, but also species that are typical of old-growth cloud forest. We suggest that these remnants may be crucial for forest birds that move across the fragmented landscape.
Keywords: Riparian strips; Bird community; Fragmented landscape; Reservoirs
Avifauna de corredores ribereños de bosque de niebla en un paisaje degradado del este de México
Resumen
Los bosques de niebla son conocidos por su notable biodiversidad y sus servicios ecosistémicos. Sin embargo, esta biodiversidad está en peligro debido a su conversión a otros usos de suelo. En su distribución más septentrional en el Neotrópico, el bosque de niebla persiste en remanentes inmersos en una matriz agrícola que aún conserva elementos arbóreos, como los corredores riparios. En este estudio caracterizamos la avifauna presente en dichos corredores en un paisaje altamente degradado de México. Clasificamos a la avifauna en términos de su estatus migratorio y de conservación, gremio trófico, masa corporal y preferencia de hábitat y estrato forestal. En 14 corredores riparios registramos 86 especies de aves. Las especies insectívoras y frugívoras representaron 79% de la riqueza total. Casi 65% de las especies prefieren los estratos forestales medios o de dosel, mientras que 46% fueron generalistas de hábitat. Aunque los corredores riparios atraviesan zonas agrícolas, siguen albergando un conjunto diverso de aves, que incluye no solo aquellas tolerantes a las perturbaciones, sino también especies características del bosque de niebla conservado. Sugerimos que estos remanentes pueden ser cruciales para las aves forestales que se desplazan por el paisaje fragmentado.
Palabras clave: Franjas riparias; Comunidad de aves; Paisaje fragmentado; Reservorios
Introduction
Tropical montane cloud forest (hereafter, cloud forest) is one of the most important terrestrial ecosystems worldwide. It provides environmental services such as carbon sequestration and water capture, and also mitigates flooding and drought (Bruijnzeel et al., 2011). Cloud forest is among the most biodiverse ecosystems in the world and hosts a remarkable diversity of flora and fauna in which spatial variation is prominent (i.e., high beta diversity), as well as a high proportion of endemic species (Aldrich et al., 2000; Karger et al., 2021). Mexican cloud forests are particularly rich in species of trees, shrubs and epiphytes, along with amphibians, reptiles, birds and mammals (Gual-Díaz & Rendón-Correa, 2014). It is estimated that Mexican cloud forests are home to 551 bird species (i.e., 50% of the total richness of the avifauna in the country), and the cloud forests in the state of Veracruz are home to 346 bird species (Navarro-Sigüenza et al., 2014). The richness of cloud forest avifauna in other parts of the country ranges from 196 to 335 species per region (Hernández-Baños et al., 1995; Navarro-Sigüenza et al., 2014). Despite its relevance, Mexican cloud forest is currently in jeopardy, not only because of agricultural expansion and uncontrolled urban growth, but also due to the illegal extraction of its species and unsustainable use of its resources (Toledo-Aceves et al., 2011). In the central part of Veracruz in the 1990s, it was estimated that there were 426 km² of cloud forest, an area that was reduced to 279 km² by 2003 (Muñoz-Villers & López-Blanco, 2008). Even though the rate of Mexican cloud forest deforestation has slowed over the last decade, today it is estimated to cover only 175 km² in Veracruz (Bonilla-Moheno & Aide, 2020). Cloud forest remnants are currently found as numerous fragments of different sizes (1 to 30 ha, usually), which are immersed in an extensive agricultural matrix (Williams-Linera et al., 2002) that still contains distinct arboreal elements such as isolated trees, living fences and forested riparian corridors. The presence of these arboreal elements within the agricultural matrix, which also include small patches (< 5 ha) of secondary forest, could be relevant to the maintenance of different groups of native flora and fauna in anthropic landscapes (Toledo-Aceves et al., 2014).
Cloud forest riparian corridors are single rows of trees growing along each side of permanent streams or rivers that have been left uncut by farmers when converting the forest to crop fields or pastures. These narrow, elongated belts or strips of tall trees have a dense woody undergrowth (riparian corridors, hereafter) and extend along rivers for kilometers. They are usually the most conspicuous arboreal element in agricultural landscapes. Forested riparian corridors that cross agricultural plots provide several environmental services, such as riverbank stabilization, nutrient recycling and enrichment, and filtering and retention of agrochemical pollutants in surface runoff, among other services (Cole et al., 2020). Additionally, riparian corridors contribute to the conservation of several taxonomic groups in anthropic landscapes, including native woody plant species (Hernández-Dávila et al., 2020), amphibians (Rodríguez-Mendoza & Pineda, 2010), bats and non-flying mammals (Griscom et al., 2007; Zarazúa-Carbajal et al., 2017). For birds, forested riparian corridors have been shown to function as refuges, foraging and nesting sites, as well as habitat corridors or stepping stones for moving across fragmented landscapes (Domínguez-López & Ortega-Álvarez, 2014; Kontsiotis et al., 2019; Lees & Peres, 2008). In lowland tropical regions that have been converted to agriculture, relatively wide riparian corridors connected to large forest remnants have been found to harbor a higher richness and abundance of forest birds than narrower and unconnected riparian corridors (Arizmendi et al., 2008; Domínguez-López & Ortega-Álvarez, 2014; Pliscoff et al., 2020). The presence of linear arboreal elements within the agricultural matrix might help to maintain bird diversity in anthropic landscapes (de Zwaan et al., 2022). To date, the majority of studies on the avifauna that uses riparian corridors in anthropic or/and fragmented landscapes has been done in lowland areas originally covered by tropical rainforest or seasonally dry tropical forest (Graham et al., 2002; Latta et al., 2012; Villaseñor-Gómez, 2008). In particular, for riparian strips of tropical montane cloud forest, Hernández-Dávila et al. (2021) found that landscape composition (i.e., urban and forest area) and vegetation structure (mean height of vegetation) positively influence the richness and abundance of generalist and specialist birds using riparian strips. However, the bird community of cloud forest riparian habitats that could potentially be using these remnants in anthropic landscapes has not been characterized to date. In this sense, taking into account that the original area covered by cloud forest has been drastically reduced by human activities and that the remaining forest fragments are surrounded by an extensive agricultural matrix, in which still there are arboreal riparian strips, we wanted to determine and characterize the composition and structure of the bird community present in these remnants. The latter, in order to find out if these riparian strips can serve as biodiversity reservoirs or landscape connectors, that facilitate the movement of birds across the landscape and which bird species use them. The objectives of this paper were: 1) to determine the richness, diversity and composition of birds that use riparian corridors of cloud forest in a highly modified anthropic landscape, and 2) to characterize the avifauna that uses these corridors in terms of their conservation and migratory status, trophic guild, and habitat preference. To date, no characterization of the bird community present in riparian corridors of cloud forest has been carried out. Given that the cloud forest is fragmented and natural or semi-natural remnants, such as arboreal riparian corridors, can support different groups of flora and fauna, it is necessary to determine and analyze the avifauna present in these corridors. This knowledge is needed to design and implement management strategies of riparian corridors and improve the odds of native cloud forest species conservation in current landscapes.
Materials and methods
The study area is located in the central part of the state of Veracruz, Mexico in the upper basin of the La Antigua River within 19°31’59”-19°22’42” N, 97°05’36”-96°57’43” W (Fig. 1). The original vegetation was cloud forest (i.e., Tropical Montane Cloud Forest) with an average annual temperature of 18 °C and total annual precipitation from 1,500 to 2,000 mm/year. Fourteen riparian corridor sites (with elevations from 1,190 up to 1,780 m asl) were selected for bird sampling. The sites selected are near to the cities of Xalapa, Coatepec, and Xico and are part of a highly fragmented landscape and are representative of the riparian corridors in the region (i.e., narrow linear bands of remnant cloud forest 2 to 5 m wide growing on both sides of permanent rivers). Since riparian corridors can be several kilometers long, we delimited each of our 14 sampling sites as a riparian tract or segment approximately 400 m long (± 16 m, s.e.) with a continuous tree canopy (i.e., uninterrupted arboreal cover). The 14 riparian segments selected were all separated by more than 1 km. Most of the segments of riparian corridors that we selected for bird sampling cross open cattle pastures, with a few of them adjoining small patches (< 4 ha) of secondary old-growth forest or different types of crop fields (mostly maize or shaded coffee plantations). Dominant tree species in the sampling sites include Platanus mexicana, Liquidambar styraciflua, Palicourea padifolia, Styrax glabrescens, and Perrottetia longistylis. The canopy of these riparian corridors is formed by tall trees usually 15 to 20 m in height, with some surpassing 30 m, however, the average tree height in the sampling sites was 6.3 (± 7.0) m. See Hernández-Dávila et al. (2020) for more details on the vegetation structure and composition of the sites sampled.
The richness and number of birds visiting each sampling site were recorded at 2 fixed point counts set along each segment at least 250 m apart (Gregory et al., 2004). Field observations were conducted in October 2017, and in January, April and July 2018, for a total of 4 visits per point over the course of 1 year, with a 75-day interval between visits to each point. For each point count, all bird sightings were recorded with binoculars over the course of 15 minutes and within 35 m distance by one of us between sunrise and 10:30 am, except on rainy days. It took 3 days to complete the 28 points of a given period. The starting point during each period was alternated to cover the whole morning schedule of observation at each point. Only birds that were perching on the woody vegetation or on the ground or riverbank were recorded. Those flying overhead or perching in open areas nearby outside the riparian corridor were not counted. Thus, a total of 112 counts (14 sites × 2 points/site × 4 visits/site) were done at 28 points, totaling 28 h of observation. In addition to the visual records, we also set mist nets to capture birds to record understory birds visiting some of the sampled corridors, but owing to time and monetary constraints, we were only able to place the nets in 6 of the 14 riparian corridor sites. These 6 sites were chosen randomly and in each of them a total of 15 nets (10 × 2.5 m, each) were set parallel to the river flow on both riversides and at least 50 m apart along the sampled segment. From August 2016 to July 2017, one of the 6 riparian corridors was selected each month for mist-netting. Nets were left in place for 3 consecutive days avoiding rainy days and opened twice a day from sunrise to 11:00 and from 16:00 to sunset. This was done at each site twice over the sampled year. Sampling effort was 1,743 h and 2,250 m² of nets (25 m²/net × 15 nets × 6 sites) in 2 sampling periods at each site. Nets were inspected by 2 people every 30 minutes or less, depending on capture intensity. Each captured bird was marked by trimming a notch at the tip of one of the tail feathers to recognize recaptured individuals. Birds were released in situ immediately after sexing, weighing, and measuring (tarsus, tail, and total length; wing chord length; beak length, width, and depth).
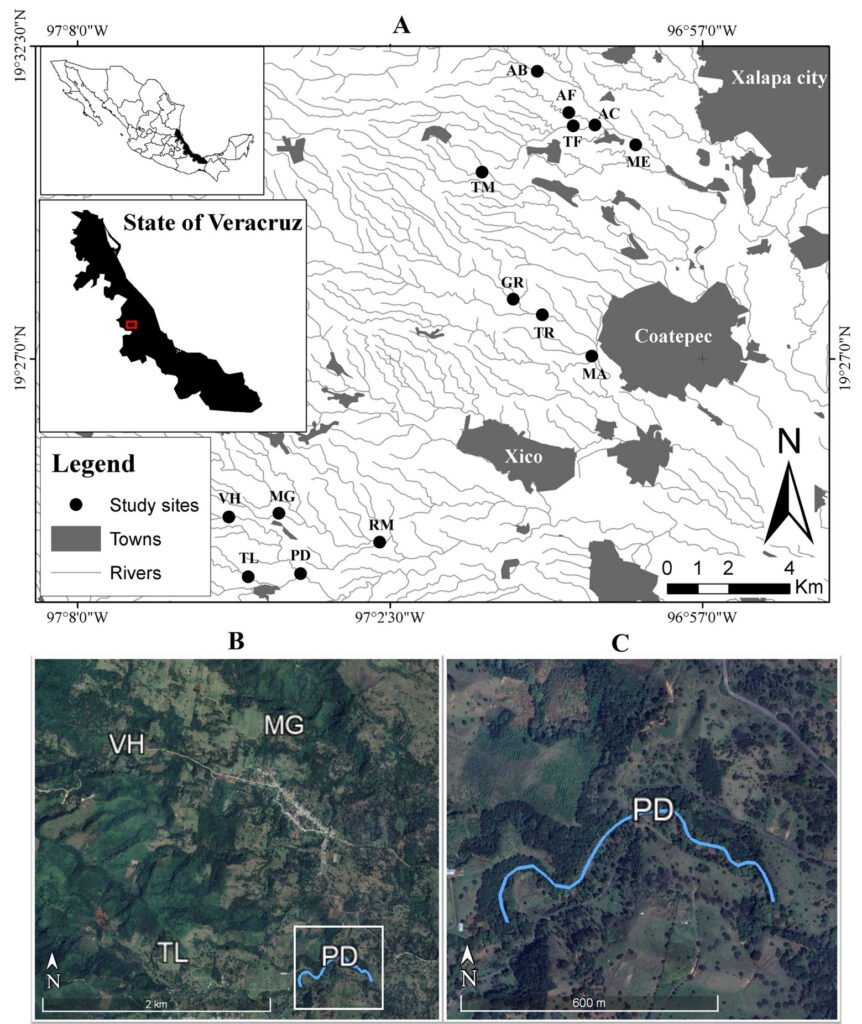
Figure 1. A, Location of the 14 segments of cloud forest riparian corridors (black circles) selected for bird sampling in central Veracruz, Mexico. The main river or streams (lines) and cities (polygons) are shown in gray. Sampling sites: Agua Bendita (AB), Agüita Fría (AF), Acuario (AC), Trucha Feliz (TF), Mariano Escobedo (ME), Truchas Martin (TM), Granada (GR), Trianon (TR), Marina (MA), Rio Matlacobatl (RM), Puente de Dios (PD), Monte Grande (MG), Tlalchy (TL) and Vista Hermosa (VH); B, image (Google Earth – Pro V 7.3.6.9345) is a close-up of 4 of the riparian corridor segments sampled (VH, MG, TL, PD); C, close-up of the PD site, with the river indicated by a blue line. Map by O. Hernández-Dávila.
Bird species were identified using the Sibley (2000) and Howell and Webb (1995) field guides. Nomenclature follows the IOC World Bird List checklist (Gill et al., 2024). Recorded birds were classified as either migratory from North America or resident species, and we also noted whether they were endemic, following Navarro-Sigüenza et al. (2007). The conservation status of each species was based on Mexican federal law NOM-059 (Semarnat 2010), and The IUCN Red List (IUCN, 2024). For trophic guild, each species was classified as: carnivore, insectivore, frugivore, granivore, nectarivore and omnivore (González-Salazar et al., 2014). Additionally, species were categorized into 3 size categories (body mass): small (< 40 g), medium (40 – 100 g) and large birds (> 100 g). Forest stratum preferences were based on Martínez-Morales (2001) and classified as: canopy species (those associated with the upper forest stratum, > 10 m above the ground); midstory species (those that preferentially use forest stratum between 5 and 10 m above the ground); midstory and canopy species (those using both midstory and canopy strata); understory species (those that frequent the forest floor up to 5 m above the ground); understory and midstory species (those using both understory and midstory strata); and all strata species (those that use all forest strata). Finally, main habitat preference was also based on Martínez-Morales (2001) and noted as: forest interior species (those which prefer forest sites away from the forest edge); forest edge species (those that preferentially use the fragment edge less than 100 m away from open areas); forest generalist species (those that are common both at the edge and the interior of forest fragments); and vegetation matrix species (those associated with the agricultural matrix or open areas). For species not reported by Martínez-Morales (2001) we did not assign categories for forest stratum preference and habitat preference. For these cases, species were classified as unknown.
The total richness recorded in riparian corridors was determined by the sum of records obtained from point records and those from mist-nets. Each bird species was characterized in relation to its migratory and conservation status, size, trophic guild, and habitat and forest stratum preferences. Due to the differences in the number of riparian corridors sampled by each sampling method (i.e., 14 for count points and 6 for mist nets) and their differences in data obtained from each method, for all the remaining analyses, only data from count points were taken into account: species accumulation curve and sample coverage were estimated to assess the sample completeness. Hill numbers were calculated to analyze the diversity of bird species in terms of effective number of species (q0), effective number of abundant species or Shannon diversity (q1) and effective number of dominant species or Simpson diversity (q2) (Hsieh et al., 2016; Jost, 2006). The rank-abundance curve of overall recorded avifauna was drawn to show graphically the structure of the bird community (Kindt & Coe, 2005). Analyses of accumulation curves, Hill numbers and range-abundance curves were performed for all riparian strips combined and separately. Finally, similarity in species composition among the 14 sampling sites was estimated using the Jaccard index distance, which varies from 0 to 1. Zero indicates that no species are shared between compared sites, and 1 indicates that species composition is identical between the sites. All analyses were run in R software using the vegan (Oksanen et al., 2020) and BiodiversityR (Kindt, 2021) packages.
Results
We recorded a total of 86 bird species in the 14 riparian corridors. These 86 species belonged to 9 orders, 27 families and 67 genera (Table 1). The richest families were Parulidae (16 species); Tyrannidae (13 spp.); Trochilidae (9 spp.) and Turdidae (6 spp.). Of the 86 bird species, there were 67 recorded at the point-counts and 48 trapped in the nets, with 29 recorded with both field methods (Table 1).
Of the 86 recorded species, 65 were resident species and 21 were migratory. Only 2 of the recorded species are endemic to Mexico: Melanotis caerulescens (Blue Mockingbird) and Cardellina rubra (Red Warbler). Four species are under special protection status by Mexican federal law (Semarnat, 2010): Accipiter striatus (Sharp-shinned Hawk), Cinclus mexicanus (American Dipper), Psarocolius montezuma (Montezuma Oropendola) and Catharus mexicanus (Black-headed Nightingale-thrush), and 1 species is threatened: Catharus frantzii (Ruddy-capped Nightingale-thrush). One species, Selasphorus rufus (Rufous Hummingbird), is regarded as Near Threatened in the IUCN list. The richest trophic guilds were insectivorous (63% of species) and frugivorous birds (15%) (Fig. 2A). Regarding forest stratum preference, 18% of the species recorded prefer the midstory to canopy strata, and 17% are midstory specialists (Fig. 2B). For habitat preference, habitat generalists were the most strongly represented accounting for 30% of richness, followed by forest-interior species with 18% (Fig. 2C). Most recorded species (66%) were relatively small with a body mass < 40 g, and only 12% of species were heavier than 100 g (Fig. 2D).
During the 28 point counts we recorded a total of 816 detections of 67 bird species using the corridors. In the mist nets placed in 6 of the corridors, we captured 273 birds belonging to 48 species. The species accumulation curve for all corridors reached a sample completeness of 97%. According to Hill numbers the bird diversity was of 67 species observed (q0), 18 common species (q1) and 8 dominant species (q2) (Fig. 3). Individually, the sample completeness for each of the 14 riparian segments varied between 79% and 93% per segment; and regarding diversity, q0 ranged from 9 to 24 bird species per segment, while q1 ranged from 5 to 13 species and q2 from 2 to 13 species (Fig. 4).
Table 1
List of the 86 bird species recorded in 14 cloud forest riparian corridors in the anthropic landscape of central Veracruz, Mexico. Shown are the number of visual detections (# detections) of each species estimated by point counts and the number of birds
captured in mist nets. Seed dispersing birds are indicated with an asterisk (*). Species are ordered by rank (only for data obtained by point counts). We also show for each species: migratory status, resident (R), North American migrant (M). Protection status: threatened (A), special protection (Pr), least concern (LC), near threatened (NT). Trophic guild: insectivore (In), frugivore (Fr), nectarivore (Ne), omnivore (Om), granivore (Gr), carnivore (Ca). Size (body mass in g): small < 40 g (1), medium-sized 40 – 100 g (2), large > 100 g (3). Habitat preference: forest interior (FI), forest edge (FE), forest generalist (FG), vegetation matrix (VM), and no information available (-). Forest stratum preference: understory (U), understory and midstory (UM), midstory (M), midstory and canopy (MC), canopy (C), and no information available (-). See below for bibliographic sources.
| Species | # detections by point counts | # captures in mist nets | Migratory status | Protection status | Trophic guild | Size category | Habitat preference | Stratum preference |
| Chlorospingus flavopectus* | 234 | 17 | R | -/LC | In-Fr | 1 | FG | MC |
| Cardellina pusilla | 102 | 15 | M | -/LC | In | 1 | FG | A |
| Psilorhinus morio* | 79 | 2 | R | -/LC | Om | 3 | FG | MC |
| Myadestes occidentalis* | 51 | 10 | R | -/LC | Fr | 2 | FG | MC |
| Empidonax difficilis | 36 | 18 | R | -/LC | In | 1 | FG | M |
| Myiozetetes similis* | 33 | 3 | R | -/LC | In-Fr | 1 | VM | C |
| Parkesia motacilla* | 20 | 14 | M | -/LC | In-Fr | 1 | FI | U |
| Psarocolius montezuma* | 20 | – | R | Pr/LC | Fr | 3 | – | – |
| Setophaga townsendi | 12 | – | M | -/LC | In | 1 | FG | MC |
| Sayornis nigricans | 11 | 2 | R | -/LC | In | 1 | – | – |
| Tityra semifasciata* | 9 | – | R | -/LC | In-Fr | 2 | – | – |
| Turdus assimilis* | 9 | 6 | R | -/LC | Fr | 2 | FE | M |
| Turdus grayi* | 9 | 6 | R | -/LC | Fr | 2 | FE | M |
| Cinclus mexicanus | 8 | – | R | Pr/LC | Ca | 2 | – | – |
| Leiothlypis ruficapilla | 8 | 1 | M | -/LC | In | 1 | FI | MC |
| Mitrephanes phaeocercus | 8 | – | R | -/LC | In | 1 | FG | M |
| Quiscalus mexicanus* | 8 | 1 | R | -/LC | Om | 3 | – | A |
| Vireo solitarius* | 8 | 3 | M | -/LC | In-Fr | 1 | FG | M |
| Henicorhina leucophrys* | 7 | 4 | R | -/LC | In-Fr | 1 | FG | U |
| Melanerpes aurifrons* | 6 | – | R | -/LC | In-Fr | 2 | – | – |
| Thraupis abbas* | 6 | 1 | R | -/LC | Fr | 2 | FE | C |
| Contopus pertinax | 5 | 1 | R | -/LC | In | 1 | FG | C |
| Cyanolyca cucullata* | 5 | – | R | -/LC | Om | 3 | FI | MC |
| Euphonia hirundinacea* | 5 | 9 | R | -/LC | Fr | 1 | – | – |
| Lepidocolaptes affinis | 5 | 3 | R | -/LC | In | 1 | FG | M |
| Mniotilta varia | 5 | 1 | M | -/LC | In | 1 | FI | MC |
| Myioborus miniatus | 5 | 1 | R | -/LC | In | 1 | FI | M |
| Chloroceryle americana | 4 | – | R | -/LC | Ca | 2 | FE | MC |
| Empidonax hammondii | 4 | – | M | -/LC | In | 1 | – | – |
| Melanerpes formicivorus* | 4 | – | R | -/LC | In-Fr | 2 | FE | MC |
| Piranga leucoptera* | 4 | – | R | -/LC | In-Fr | 1 | – | – |
| Table 1. Continued | ||||||||
| Species | # detections by point counts | # captures in mist nets | Migratory status | Protection status | Trophic guild | Size category | Habitat preference | Stratum preference |
| Catharus frantzii* | 3 | 4 | R | A/LC | Fr | 1 | FI | UM |
| Momotus coeruliceps* | 3 | 2 | R | -/LC | In-Fr | 3 | FG | M |
| Myiarchus tuberculifer* | 3 | 1 | R | -/LC | In-Fr | 1 | FG | M |
| Polioptila caerulea | 3 | – | M | -/LC | In | 1 | FG | MC |
| Ptiliogonys cinereus* | 3 | – | R | -/LC | In-Fr | 1 | FG | C |
| Rupornis magnirostris | 3 | 1 | R | -/LC | Ca | 3 | – | – |
| Vireo cassini* | 3 | – | M | -/LC | In-Fr | 1 | – | – |
| Amazona albifrons | 2 | – | R | -/LC | Gr | 3 | – | – |
| Basileuterus belli* | 2 | 11 | R | -/LC | In-Fr | 1 | FI | UM |
| Basileuterus rufifrons | 2 | – | R | -/LC | In | 1 | FG | UM |
| Chlorophonia elegantissima * | 2 | – | R | -/LC | Fr | 1 | FI | MC |
| Dendrocincla homochroa | 2 | 1 | R | -/LC | In | 2 | – | – |
| Leptotila verreauxi | 2 | – | R | -/LC | Gr | 3 | FG | U |
| Pyrocephalus rubinus | 2 | – | R | -/LC | In | 1 | – | – |
| Saltator atriceps | 2 | 2 | R | -/LC | Fr | 2 | – | – |
| Setophaga tigrina | 2 | – | M | -/LC | In | 1 | – | – |
| Setophaga virens | 2 | – | M | -/LC | In | 1 | FI | MC |
| Sporophila morelleti | 2 | 1 | R | -/LC | Gr | 1 | – | – |
| Campylorhynchus zonatus | 1 | – | R | -/LC | In | 2 | – | – |
| Cardellina rubra | 1 | – | R | -/LC | In | 1 | – | – |
| Catharus mexicanus* | 1 | 12 | R | Pr/LC | Fr | 1 | FG | U |
| Corthylio calendula | 1 | – | M | -/LC | In | 1 | FG | U |
| Dives dives* | 1 | 2 | R | -/LC | In-Fr | 2 | VM | A |
| Dumetella carolinensis* | 1 | 2 | M | -/LC | In-Fr | 2 | VM | M |
| Empidonax minimus | 1 | – | M | -/LC | In | 1 | – | – |
| Icterus bullockii* | 1 | – | R | -/LC | In-Fr | 1 | FE | M |
| Megarynchus pitangua* | 1 | – | R | -/LC | In-Fr | 2 | – | – |
| Ortalis vetula* | 1 | – | R | -/LC | Fr | 3 | FE | M |
| Pachyramphus aglaiae | 1 | – | R | -/LC | In | 1 | FI | MC |
| Piaya cayana* | 1 | – | R | -/LC | In-Fr | 3 | FG | M |
| Piranga flava* | 1 | – | R | -/LC | In-Fr | 1 | FE | MC |
| Piranga rubra* | 1 | – | M | -/LC | In-Fr | 1 | FI | MC |
| Seiurus aurocapilla | 1 | 1 | M | -/LC | In | 1 | FI | U |
| Setophaga nigrescens | 1 | – | M | -/LC | In | 1 | – | – |
| Setophaga ruticilla | 1 | – | M | -/LC | In | 1 | – | – |
| Tyrannus melancholicus | 1 | – | R | -/LC | In | 1 | VM | C |
| Accipiter striatus | – | 1 | R | Pr/LC | Ca | 3 | – | – |
| Archilochus colubris | – | 1 | M | -/LC | Ne | 1 | FI | UM |
| Table 1. Continued | ||||||||
| Species | # detections by point counts | # captures in mist nets | Migratory status | Protection status | Trophic guild | Size category | Habitat preference | Stratum preference |
| Arremon brunneinucha* | – | 14 | R | -/LC | In-Fr | 2 | FG | M |
| Basileuterus culicivorus | – | 2 | R | -/LC | In | 1 | FG | UM |
| Campylopterus hemileucurus | – | 20 | R | -/LC | Ne | 1 | – | – |
| Cardellina canadensis | – | 1 | M | -/LC | In | 1 | FI | U |
| Catharus aurantiirostris* | – | 2 | R | -/LC | Fr | 1 | FG | U |
| Chlorestes candida | – | 1 | R | -/LC | Ne | 1 | – | – |
| Eugenes fulgens | – | 2 | R | -/LC | Ne | 1 | FE | M |
| Lampornis amethystinus | – | 21 | R | -/LC | Ne | 1 | FG | U |
| Melanotis caerulescens* | – | 1 | R | -/LC | In-Fr | 2 | FE | U |
| Myiodynastes luteiventris* | – | 2 | R | -/LC | In-Fr | 2 | FI | MC |
| Pampa curvipennis | – | 12 | R | -/LC | Ne | 1 | VM | UM |
| Pitangus sulphuratus* | – | 1 | R | -/LC | In-Fr | 2 | FG | A |
| Saucerottia beryllina | – | 12 | R | -/LC | Ne | 1 | – | – |
| Saucerottia cyanocephala | – | 19 | R | -/LC | Ne | 1 | FE | UM |
| Selasphorus rufus | – | 1 | M | -/NT | Ne | 1 | – | – |
| Stelgidopteryx serripennis | – | 1 | R | -/LC | In | 1 | FG | C |
| Vireo gilvus* | – | 1 | R | -/LC | In-Fr | 1 | FI | C |
*Seed dispersing birds: based on field data from Hernández-Dávila et al. (2022) and Hernández-Ladrón De Guevara et al. (2012). Migratory status: from Howell and Webb (1995) and Sibley (2000). Size: from Martínez-Morales (2001), Sibley (2000), and birds captured in mist nets (this study). Habitat preference and stratum preference: from Martínez-Morales (2001).
The most common species recorded visually was Chlorospingus flavopectus (Common Chlorospingus) with 234 detections (Fig. 5A), followed by Cardellina pusilla (Wilson’s Warbler) with 102, Psilorhinus morio (Brown Jay) with 79 and Myadestes occidentalis (Brown-backed Solitaire) with 51 detections (see species detection data in Table 1). These 4 dominant species accounted for 45% of total detections recorded in the point counts. There were 10 species with only 2 detections (i.e., doubletons) and 18 species with only 1 (singletons), and these 28 extremely rare species accounted for less than 5% of total detections and 42% of the 67 species recorded in the point counts. In general, the pattern of dominance by the 4 species mentioned occurred in each riparian corridor sampled, concentrating most of the bird detections at each site, with several species having much fewer detections per site (Fig. 5B).
Bird species richness per site estimated in point counts varied from 9 (TM site) to 33 species (MA site), with 10 of the 14 segments sampled having fewer than 20 species each. Similarity between sites was low (Jaccard index, J < 0.4) for most of the paired comparisons (Table 2), with the highest level of similarity between the TL and AB sites (0.53) and the lowest between MA and AF (0.10).
Discussion
In general, and pooling all sampled riparian corridors, the sample completeness was high (97%), recording a total richness of 86 bird species in the 14 segments sampled in the fragmented cloud forest landscape of central Veracruz, Mexico. This richness is comparable to that reported in similar studies carried out in relatively large (> 3 ha) remnant fragments of cloud forest in the same region, where up to 75-100 bird species have been detected (Rueda-Hernández et al., 2015; Serna-Lagunes et al., 2023). These 14 riparian corridors harbor 24% of the bird richness reported for cloud forest throughout the entire state of Veracruz (Navarro-Sigüenza et al., 2014). The richness recorded in our study represents between 25 and 43% of the avifauna reported in other regions of Mexico with cloud forest, where 196 to 335 bird species have been found (Martínez-Morales, 2007; Navarro-Sigüenza et al., 2014). The richness detected suggests that riparian corridors are important elements in deforested landscapes of cloud forest for numerous birds. Other studies in different sites have shown that these elements of the landscape can serve as refuges, foraging areas and even as reproductive (nesting) sites (Hawes et al., 2008; de Zwaan et al., 2022), as well as making it possible for birds to move across large open areas in agricultural landscapes (Gillies & St. Clair, 2010; Pliscoff et al., 2020). Both resident species and migratory birds, visit and use forested riparian corridors during their autumn-winter stay in the tropics (Skagen et al., 1998; Villaseñor-Gómez, 2008). As shown in our results: 24% of the recorded species were North American migratory species. The migratory bird Cardellina pusilla, abundant in cloud forest as well as in shaded coffee plantations in Veracruz (Navarro-Sigüenza et al., 2014), was the second most common species in our study. By far, the most dominant species in our study was Chlorospingus flavopectus, a resident bird regarded as a forest generalist that is common to old-growth forest, forest edge habitats and patches of secondary forest (Cruz-Angón et al., 2008; Martínez-Morales, 2007; Renner et al., 2006). Myadestes occidentalis was the fourth most dominant species in riparian corridors, which is considered a typical cloud forest species (Caballero-Cruz et al., 2020). This pattern of dominance is similar to that recorded by Martínez-Morales (2001) who reported C. flavopectus, M. occidentalis, Henicorhina leucophrys, Catharus mexicanus, and Trogon mexicanus as the most dominant species in conserved fragments of cloud forest. Except for T. mexicanus, the mentioned species were recorded in this study. Another very common bird in our study was Psilorhinus morio, a habitat generalist associated with disturbed areas, and common in small forest fragments of cloud forest (Serna-Lagunes et al., 2023), in rainforest riparian corridors that cross pastures (Graham et al., 2002), as well as in open agricultural areas with scant arboreal cover (Cerezo et al., 2009). It is important to mention that P. morio is a large species (> 100 gr), only 12% of the species recorded belong to this size category, while 65% are small species (< 40 gr). The conversion of forest for agricultural purposes mainly affects the presence of large bird species due to the reduction in food availability as well as fewer nesting and roosting sites (Gomes et al., 2008; Martínez-Morales, 2001). Thus, large bird species are the most strongly affected by the reduction of forest cover, while small birds are more vagile and tolerant to deforestation, explaining the predominance of species smaller than 40 g in the riparian corridors studied.
Table 2
Similarity distance in bird species composition (Jaccard index) among the 14 riparian corridors sampled (upper-right side of Table) with point counts (see Materials and methods), showing the number of species shared between riparian corridors (lower-left side), and the total number of species in each corridor (diagonal black cells). Gray-shaded cells highlight the highest and lowest values of similarity distance. The names of riparian corridors sampled are abbreviated as in Figure 1.
| AC | AB | AF | TR | GR | MA | ME | MG | PD | RM | TL | TF | TM | VH | |
| AC | 21 | 0.36 | 0.39 | 0.39 | 0.37 | 0.26 | 0.31 | 0.37 | 0.43 | 0.29 | 0.26 | 0.34 | 0.30 | 0.27 |
| AB | 9 | 13 | 0.33 | 0.35 | 0.43 | 0.24 | 0.30 | 0.38 | 0.39 | 0.32 | 0.53 | 0.35 | 0.38 | 0.32 |
| AF | 9 | 6 | 11 | 0.21 | 0.24 | 0.10 | 0.14 | 0.29 | 0.36 | 0.17 | 0.33 | 0.26 | 0.43 | 0.28 |
| TR | 11 | 8 | 5 | 18 | 0.52 | 0.24 | 0.29 | 0.21 | 0.32 | 0.27 | 0.19 | 0.29 | 0.23 | 0.25 |
| GR | 11 | 10 | 6 | 13 | 20 | 0.33 | 0.32 | 0.24 | 0.39 | 0.33 | 0.27 | 0.36 | 0.32 | 0.33 |
| MA | 11 | 9 | 4 | 10 | 13 | 33 | 0.31 | 0.17 | 0.27 | 0.43 | 0.21 | 0.28 | 0.17 | 0.25 |
| ME | 9 | 6 | 3 | 7 | 8 | 11 | 13 | 0.21 | 0.33 | 0.37 | 0.18 | 0.35 | 0.29 | 0.25 |
| MG | 10 | 8 | 6 | 6 | 7 | 7 | 5 | 16 | 0.35 | 0.25 | 0.32 | 0.42 | 0.32 | 0.27 |
| PD | 12 | 9 | 8 | 9 | 11 | 11 | 8 | 9 | 19 | 0.30 | 0.33 | 0.48 | 0.33 | 0.35 |
| RM | 10 | 9 | 5 | 9 | 11 | 17 | 10 | 8 | 8 | 24 | 0.32 | 0.40 | 0.22 | 0.24 |
| TL | 7 | 9 | 6 | 5 | 7 | 8 | 4 | 7 | 7 | 9 | 13 | 0.29 | 0.29 | 0.25 |
| TF | 10 | 8 | 6 | 8 | 10 | 11 | 8 | 10 | 12 | 12 | 7 | 18 | 0.35 | 0.43 |
| TM | 7 | 6 | 6 | 5 | 7 | 6 | 5 | 6 | 8 | 6 | 5 | 7 | 9 | 0.40 |
| VH | 7 | 6 | 5 | 6 | 8 | 9 | 5 | 6 | 10 | 7 | 5 | 9 | 6 | 12 |
Figure 2. Characterization of the avifauna recorded in 14 segments of cloud forest riparian corridors sampled in central Veracruz, Mexico. A, Trophic guild: insectivore (In), frugivore (Fr), nectarivore (Ne), carnivore (Ca), omnivore (Om), and granivore (Gr); B, forest stratum preference: canopy (C), midstory and canopy (MC), midstory (M), understory and midstory (UM), understory (U), and all strata (A); C, habitat preference: forest generalist (FG), forest interior (FI), forest edge (FE), and vegetation matrix (VM); D, size (body mass): small (1 – 40 g), medium (40 – 100 g) and large (100 – 500 g).
Insectivorous birds were the richest and most abundant trophic guild in the riparian corridors sampled and are also the most common guild in intact cloud forest (Martínez-Morales, 2007). Frugivorous birds were the second richest guild and were relatively abundant in our riparian corridors. These species, together with omnivorous species, are particularly important in forest regeneration due to their role as seed dispersers of forest plants. Some of the most important frugivores that are efficient dispersers of cloud forest trees, shrubs and other zoochorous plants include M. occidentalis, C. flavopectus, P. morio, and several species of the genera Turdus, Catharus, and Euphonia (Hernández-Dávila et al., 2022; Hernández-Ladrón De Guevara et al., 2012), all of which were recorded in our study. Seed dispersal by birds is crucial for cloud forest restoration since most plant species native to this forest depend on vertebrate frugivores for dispersal (Jordano et al., 2011). Forest frugivores usually avoid open areas that are devoid of perching sites, and this is one of the strongest limitations to forest restoration due to the limited or absent immigration of woody plant seeds into agricultural areas (Holl et al., 2000). However, it has been recorded that the density of linear forest or wooded patches such as riparian strips and live fences can increase bird diversity in agricultural landscapes (Wilson et al., 2017). In this sense cloud forest riparian corridors could contribute to species movement across the landscape. Thus, forested riparian corridors within the agricultural matrix facilitate that frugivorous birds will visit these disturbed sites and disperse seeds across and into the site. Another group of birds that is crucial for plant reproduction are the nectarivorous species, with several species of hummingbirds being particularly important. Of the 26 hummingbird species reported for Mexican cloud forest (Navarro-Sigüenza et al., 2014), 9 were recorded in the riparian corridors that we studied. The presence of birds that are seed or pollen vectors in riparian corridors contributes greatly to connectivity in anthropic landscapes and are essential to biodiversity conservation and forest regeneration and restoration in these landscapes.
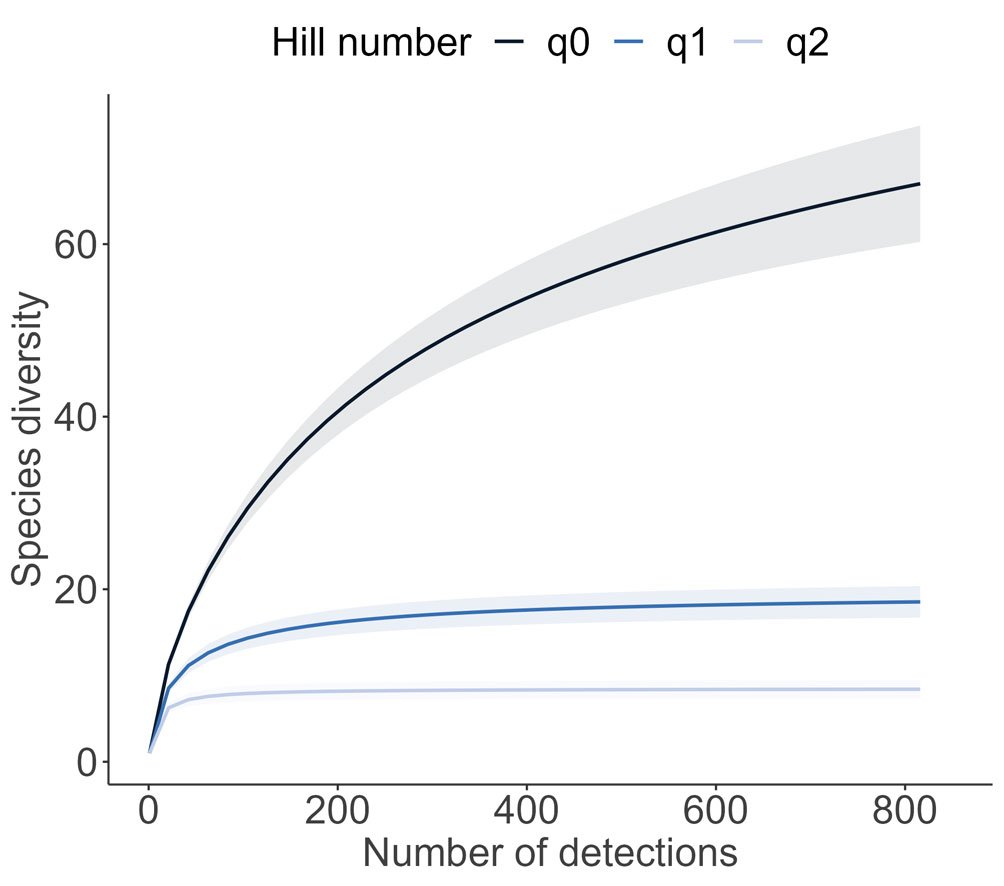
Figure 3. Species accumulation curve and Hill numbers of the bird community recorded in 14 cloud forest riparian corridors. Shaded area delimits 95% confidence intervals.
As expected by the high degree of anthropic disturbance in the landscape we studied, the richest groups of birds in the cloud forest riparian corridors were forest generalist species and those associated with the vegetation of the agricultural matrix (sensu Martínez-Morales, 2001), which together represented 53% of the avifauna we recorded. The presence and wide distribution of different arboreal elements within the current anthropic landscape could explain the presence of forest interior bird species in riparian corridors. Changes in the structure and floristic composition of the original vegetation of a given site, resulting from human activities also lead to changes in the community attributes of the avifauna (Martínez-Morales, 2005). Among the most salient changes in the forested riparian corridors that cross agricultural areas is the high abundance of plants that are common in large canopy gaps or associated with disturbed sites, including several species of the families Piperaceae, Melastomataceae, and Rubiaceae, which were common in our study (Hernández-Dávila et al., 2020). These vegetation changes are more favorable to habitat generalists than to bird species associated with the interior of large forest fragments, explaining the lower proportion of species whose preferred habitat is the forest interior in our results. Studies in different regions report an increase in the richness and abundance of habitat generalist birds in forest edge habitats, where forest interior birds decrease (de Zwaan et al., 2022; Watson et al., 2004; Wilson et al., 2017).
For cloud forest, Martínez-Morales (2005) found that fragment size positively affects the richness and abundance of both generalist and forest-interior birds. In addition, for riparian corridors of cloud forest, Hernández-Davila et al. (2021) found that the richness and abundance of generalist and specialist birds showed a differential response to the amount of forest and urban cover in the vicinity of riparian strips. The percentage of urban cover near the riparian strip negatively affected the abundance of forest interior species and positively affected generalist species, whereas surprisingly the amount of forest area nearby the strip does not seem to influence the richness and abundance of birds using the riparian strip. These results could explain the differences between the number of generalist and interior species found in our study, as well as the differences of the Hill diversity values among the sampled corridors. Although this study did not analyze explicitly aspects of landscape configuration, it is relevant to mention that, although the riparian corridors are narrow remnants of just a few meters wide (< 10 m), both generalist and forest interior species were recorded in them. This suggests that these remnants may harbor a wealth of bird species regardless of their habitat preference. In fact, 2 of the bird species recorded are endemic to Mexico, another 4 are protected by law and 1 is threatened. This highlights the importance of riparian corridors as reservoirs of birds within anthropic landscapes, particularly species native to the cloud forest including resident and migratory birds. It is important to note that 3 of the species recorded in riparian corridors; Quiscalus mexicanus, Rupornis magnirostris, and Pyrocephalus rubinus could be regarded as urban birds (Maya-Elizarrarás, 2011; Ruelas & Aguilar, 2010).
As far as we know, this study is the first to describe and characterize the avifauna present in riparian corridors of the threatened cloud forest, however, it is important to take into account that, despite the fact that our point counts had a sufficient separation and elapsed time between observations to warrant independent detections and no overflying individuals or auditory recordings were included, we might have overestimated species abundances, in particular because birds move frequently along the forest strips (personal observation, OHD). Also, because mist nets capture understory species more frequently, species richness of mid- to high- strata birds are usually underestimated with nets. This study focused on riparian corridors and did not include other types of natural remnants or conserved forest fragments present in the region and part of the current mosaic of the anthropic fragmented landscape. Therefore, more studies are needed to determine the importance of riparian corridors for conserving bird diversity in comparison with other natural remnants of cloud forest.
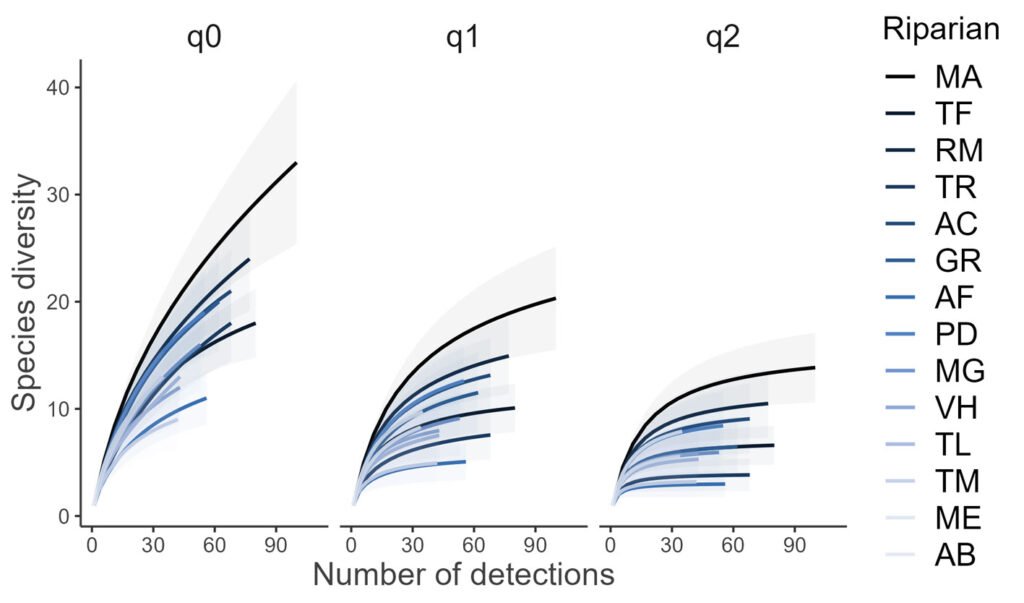
Figure 4. Species accumulation curve and Hill numbers (q0, q1, and q2) of birds recorded in each riparian corridor. Sites are ordered according to richness; dark blue curves correspond to the riparian corridors with the highest richness (e.g., MA), while light blue curves correspond to the sites with the lowest richness (e.g., AB). Shaded area delimits 95% confidence intervals. Abbreviations of riparian corridors can be found in Figure 1.
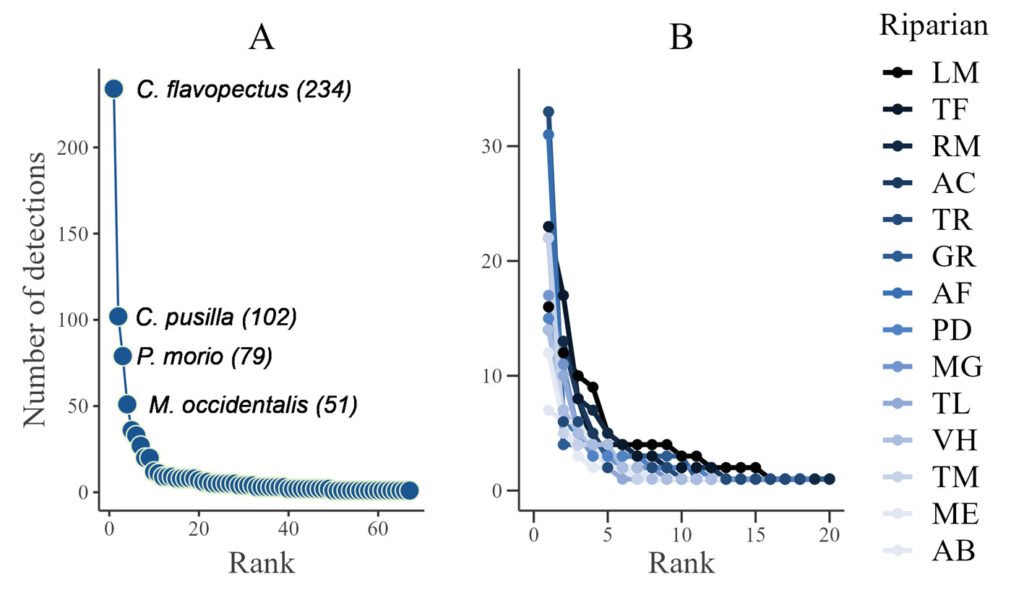
Figure 5. A, Rank-abundance curve for the avifauna recorded in 14 segments of cloud forest riparian corridors in central Veracruz, Mexico. The 4 most common species were: C. flavopectus, C. pusilla, P. morio, and M. occidentalis, the number of total detections is given in parenthesis; B, Rank-abundance curve in each riparian corridor. Sites are ordered according to number of detections; dark blue curves correspond to the riparian corridors with the highest number of detections (e.g., LM), while light blue curves correspond to the sites with the lowest number of detections (e.g., AB). Abbreviations of riparian corridors can be found in Figure 1.
In conclusion, our results show that the cloud forest riparian corridors that cross open agricultural areas harbor a diverse assemblage of bird species, which includes not only those tolerant of or associated with disturbance, but also species that are typical of intact patches of old-growth cloud forest. These would be absent in areas that are completely devoid of trees. Additionally, several of the birds recorded in our study are effective seed dispersers of cloud forest plants. For current deforested landscapes, this strongly suggests that cloud forest riparian corridors could be key elements in forest restoration efforts and for the conservation of bird biodiversity in agricultural landscapes. Landscape management plans designed to encourage the permanence and sustainable management of these forested corridors within the agricultural matrix will not only help in the conservation of the avifauna, but also in the restoration of degraded landscapes, thanks to the ecosystem services provided by birds, including the pollination and seed dispersal of plants native to the cloud forest.
Acknowledgement
We are grateful to María de los Ángeles García and Diana Vázquez for their valuable help in the field. The Instituto de Ecología, A.C. and Idea Wild provided the space and equipment that made this study possible. Bianca Delfosse translated the text from the original in Spanish and edited subsequent versions of the manuscript. We also thank two anonymous reviewers for their suggestions to improve the manuscript. This work was supported by The Rufford Foundation (grant number 20471-1 to OHD) and the Consejo Nacional de Ciencia y Tecnología (grant numbers CONACYT-CB-2016-01 to VJS, and graduate scholarship CONACYT-285962 to OHD).
References
Aldrich, M., Bubb, P., Hostettler, S., & van de Wiel, H. (2000). Tropical montane cloud forests: time for action. WWF International/IUCN The World Conservation Union.
Arizmendi, M. D. C., Dávila, P., Estrada, A., Figueroa, E., Márquez-Valdelamar, L., Lira, R. et al. (2008). Riparian Mesquite bushes are important for bird conservation in tropical arid Mexico. Journal of Arid Environments, 72, 1146–1163. https://doi.org/https://doi.org/10.1016/j.jaridenv.2007.12.017
Bonilla-Moheno, M., & Aide, T. M. (2020). Beyond deforestation: Land cover transitions in Mexico. Agricultural Systems, 178, 102734. https://doi.org/https://doi.org/10.1016/j.agsy.2019.102734
Bruijnzeel, L. A., Scatena, F. N., & Hamilton, L. S. (2011). Tropical montane cloud forests. Science for conservation and management (International Hydrology Series). Cambridge University Press.
Caballero-Cruz, P., Vargas-Noguez, G., & Ortiz-Pulido, R. (2020). Especies de aves en riesgo en el bosque mesófilo de montaña en cinco AICA de la sierra Madre Oriental, México. Huitzil, Revista Mexicana de Ornitología, 21, e-490. https://doi.org/https://doi.org/10.28947/hrmo.2020.21.1.472
Cerezo, A., Robbins, C. S., & Dowell, B. (2009). Uso de hábitats modificados por aves dependientes de bosque tropical en la región caribeña de Guatemala. Revista de Biología Tropical, 57, 401–419. https://doi.org/https://doi.org/10.15517/rbt.v57i1-2.11355
Cole, L. J., Stockan, J., & Helliwell, R. (2020). Managing riparian buffer strips to optimise ecosystem services: A review. Agriculture, Ecosystems and Environment, 296, 106891. https://doi.org/https://doi.org/10.1016/j.agee.2020.106891
Cruz-Angón, A., Scott, S. T., & Greenberg, R. (2008). An experimental study of habitat selection by birds in a coffee plantation. Ecology, 89, 921–927. https://doi.org/https://doi.org/10.1890/07-0164.1
de Zwaan, D. R., Alavi, N., Mitchell, G. W., Lapen, D. R., Duffe, J., & Wilson, S. (2022). Balancing conservation priorities for grassland and forest specialist bird communities in agriculturally dominated landscapes. Biological Conser-
vation, 265, 109402. https://doi.org/10.1016/j.biocon.2021.109402
Domínguez-López, M. E., & Ortega-Álvarez, R. (2014). The importance of riparian habitats for avian communities in a highly human-modified Neotropical landscape. Revista Mexicana de Biodiversidad, 85, 1217–1227. https://doi.org/10.7550/rmb.43849
Gill, F., Donsker, D., & Rasmussen, P. (2024). IOC World Bird List (v14.1). https://www.worldbirdnames.org/new/
Gillies, C. S., & St. Clair, C. C. (2010). Functional responses in habitat selection by tropical birds moving through fragmented forest. Journal of Applied Ecology, 47, 182–190. https://doi.org/10.1111/j.1365-2664.2009.01756.x
Gomes, G. L. L., Oostra, V., Nijman, V., Cleef, A. M., & Kappelle, M. (2008). Tolerance of frugivorous birds to habitat disturbance in a tropical cloud forest. Biological Conservation, 141, 860–871. https://doi.org/10.1016/j.biocon.2008.01.007
González-Salazar, C., Martínez-Meyer, E., & López-Santiago, G. (2014). A hierarchical classification of trophic guilds for North American birds and mammals. Revista Mexicana de Biodiversidad, 85, 931–941. https://doi.org/10.7550/rmb.38023
Graham, C., Martínez-Leyva, J. E., & Cruz-Paredes, L. (2002). Use of fruiting trees by birds in continuous forest and riparian forest remnants in Los Tuxtlas, Veracruz, Mexico. Biotropica, 34, 589–597. https://doi.org/10.1111/j.1744-7429.2002.tb00578.x
Gregory, R. D., Gibbons, D. W., & Donald, P. F. (2004). Bird census and survey techniques. In W. J. Sutherland, I. Newton, and R. Green (Eds.), Bird ecology and conservation: a handbook of techniques (pp. 17–56). Oxford: Oxford University Press.
Griscom, H. P., Kalko, E. K., & Ashton, M. S. (2007). Frugivory by small vertebrates within a deforested, dry tropical region of Central America. Biotropica, 39, 278–282. https://doi.org/10.1111/j.1744-7429.2006.00242.x
Gual-Díaz, M., & Rendón-Correa, A. (2014). Bosques mesófilos de montaña de México: diversidad, ecología y manejo. México D.F.: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad.
Hawes, J., Barlow, J., Gardner, T. A., & Peres, C. A. (2008). The value of forest strips for understorey birds in an Amazonian plantation landscape. Biological Conservation, 141, 2262–2278. https://doi.org/10.1016/j.biocon.2008.06.017
Hernández-Baños, B. E., Peterson, A. T., Navarro-Sigüenza, A. G., & Escalante-Pliego, P. B. (1995). Bird faunas of the humid montane forests of Mesoamerica: biogeographic patterns and priorities for conservation. Bird Conservation International, 5, 251–277. https://doi.org/10.1017/S0959270900001039
Hernández-Dávila, O. A., Laborde, J., Sosa, V. J., & Díaz-Castelazo, C. (2022). Interaction network between frugivorous birds and zoochorous plants in cloud forest riparian strips immersed in anthropic landscapes. Avian Research, 13, 100046. https://doi.org/10.1016/j.avrs.2022.100046
Hernández-Dávila, O. A., Laborde, J., Sosa, V. J., Gallardo-Hernández, C., & Díaz-Castelazo, C. (2020). Forested riparian belts as reservoirs of plant species in fragmented landscapes of tropical mountain cloud forest. Botanical Sciences, 98, 288–304. https://doi.org/https://doi.org/10.17129/botsci.2497
Hernández-Dávila, O. A., Sosa, V. J., & Laborde, J. (2021). Effects of landscape context and vegetation attributes on understorey bird communities of cloud forest riparian belts. Ecological Engineering, 167, 106269. https://doi.org/10.1016/j.ecoleng.2021.106269
Hernández-Ladrón De Guevara, I., Rojas-Soto, O. R., López-Barrera, F., Puebla-Olivares, F., & Díaz-Castelazo, C. (2012). Dispersión de semillas por aves en un paisaje de bosque mesófilo en el centro de Veracruz, México: Su papel en la restauración pasiva. Revista Chilena de Historia Natural, 85, 89–100. https://doi.org/10.4067/S0716-078X2012000100007
Holl, K. D., Loik, M. E., Lin, E. H. V., & Samuels, I. A. (2000). Tropical montane forest restoration in Costa Rica: overcoming barriers to dispersal and establishment. Restoration Ecology, 8, 339–349. https://doi.org/https://doi.org/10.1046/j.1526-100x.2000.80049.x
Howell, N. G., & Webb, S. (1995). A guide to the birds of Mexico and Northern Central America. Oxford: Oxford University Press.
Hsieh, T. C., Ma, K. H., & Chao, A. (2016). iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods in Ecology and Evolution, 7, 1451–1456. https://doi.org/10.1111/2041-210X.12613
IUCN (08 de junio de 2024). The IUCN Red List of Threatened Species. Version 2024-1. https://www.iucnredlist.org
Jordano, P., Pierre-Michel, F., Lambert, J. E., Böhning-Gaese, K., Traveset, A., & Wright, S. J. (2011). Frugivores and seed dispersal: mechanisms and consequences for biodiversity of a key ecological interaction. Biology Letters, 7, 321–323. https://doi.org/https://doi.org/10.1098/rsbl.2010.0986
Jost, L. (2006). Entropy and diversity. Oikos, 113, 363–375. https://doi.org/https://doi.org/10.1111/j.2006.0030-1299.14714.x
Karger, D. N., Kessler, M., Lehnert, M., & Jetz, W. (2021). Limited protection and ongoing loss of tropical cloud forest biodiversity and ecosystems worldwide. Nature Ecology and Evolution, 5, 854–862. https://doi.org/10.1038/s41559-021-01450-y
Kindt, R. (2021). BiodiversityR. Package for Community Ecology and Suitability Analysis (2.16-1). http://www.worldagroforestry.org/output/tree-diversity-analysis
Kindt, R., & Coe, R. (2005). Tree diversity analysis. A manual and software for common statistical methods for ecological and biodivserity studies. Nairobi: World Agroforestry Centre (ICRAF).
Kontsiotis, V., Zaimes, G. N., Tsiftsis, S., Kiourtziadis, P., & Bakaloudis, D. (2019). Assessing the influence of riparian vegetation structure on bird communities in agricultural Mediterranean landscapes. Agroforestry Systems, 93, 675–687. https://doi.org/10.1007/s10457-017-0162-x
Latta, S. C., De La Cueva, H., & Harper, A. B. (2012). Abundance and site fidelity of migratory birds wintering in riparian habitat of Baja California. Western Birds, 43, 90–101.
Lees, A. C., & Peres, C. A. (2008). Conservation value of remnant riparian forest corridors of varying quality for amazonian birds and mammals. Conservation Biology, 22, 439–449. https://doi.org/10.1111/j.1523-1739.2007.00870.x
Martínez-Morales, M. A. (2001). Forest fragmentation effects on bird communities of tropical montane cloud forests in eastern Mexico (Ph.D. Thesis). Cambridge UK: University of Cambridge.
Martínez-Morales, M. A. (2005). Landscape patterns influencing bird assemblages in a fragmented Neotropical cloud forest. Biological Conservation, 121, 117–126. https://doi.org/10.1016/j.biocon.2004.04.015
Martínez-Morales, M. A. (2007). Avifauna del bosque mesófilo de montaña del noreste de Hidalgo, México. Revista Mexicana de Biodiversidad, 78, 149–162. https://doi.org/https://revista.ib.unam.mx/index.php/bio/article/view/395
Maya-Elizarrarás, E. (2011). Aves explotadoras de áreas verdes urbanas: un ejemplo de la zona metropolitana de Guadalajara, Jalisco. El Canto Del Cenzontle, 2, 104–109.
Muñoz-Villers, L. E., & López-Blanco, J. (2008). Land use/cover changes using Landsat TM/ETM images in a tropical and biodiverse mountainous area of central-eastern Mexico. International Journal of Remote Sensing, 29, 71–93. https://doi.org/10.1080/01431160701280967
Navarro-Sigüenza, A. G., Lira-Noriega, A., Peterson, A. T., Oliveras de Ita, A., & Gordillo-Martínez, A. (2007). Diversidad, endemismo y conservación de las aves. In I. Luna, J. J. Morrone, & D. Espinosa (Eds.), Biodiversidad de la Faja Volcánica Transmexicana (pp. 461–483). México D.F.: Universidad Nacional Autónoma de México.
Navarro-Sigüenza, A. G., Gómez de Silva, H., Gual-Díaz, M., Sánchez-Gonzales, L. A., & Pérez-Villafaña, M. (2014). La importancia de las aves del bosque mesófilo de montaña de México In M. Gual-Díaz, & A. Rendón-Correa (Eds.), Bosques mesófilos de montaña de México: diversidad, ecología y manejo (pp. 279–299). Conabio.
Oksanen, J., Guillaume, B. F., Kindt, R., Legendre, P., McGlinn, D., Minchin, P. R. et al. (2020). Community Ecology Package (2.5-7). http://mirror.bjtu.edu.cn/cran/web/packages/vegan/vegan.pdf
Pliscoff, P., Simonetti, J. A., Grez, A. A., Vergara, P. M., & Barahona-Segovia, R. M. (2020). Defining corridors for movement of multiple species in a forest-plantation landscape. Global Ecology and Conservation, 23, e011108. https://doi.org/10.1016/j.gecco.2020.e01108
Renner, S. C., Walterrt, M., & Mühllenberg, M. (2006). Comparison of bird communities in primary vs. young secondary tropical montane cloud forest in Guatemala. Biodiversity and Conservation, 15, 1545–1575. https://doi.org/10.1007/s10531-005-2930-6
Rodríguez-Mendoza, C., & Pineda, E. (2010). Importance of riparian remnants for frog species diversity in a highly fragmented rainforest. Conservation Biology, 6, 781–784. https://doi.org/10.1098/rsbl.2010.0334
Rueda-Hernández, R., MacGregor-Fors, I., & Renton, K. (2015). Shifts in resident bird communities associated with cloud forset patch size in Central Veracruz, Mexico. Avian Conservation and Ecology, 10. https://doi.org/http://dx.doi.org/10.5751/ACE-00751-100202
Ruelas, I. E., & Aguilar, R. S. H. (2010). La avifauna urbana del parque ecológico Macuiltépetl en Xalapa. Ornitología Neotropical, 21, 87–103.
Semarnat (Secretaría del Medio Ambiente y Recursos Naturales). (2010). Norma Oficial Mexicana NOM-059-SEMARNAT-2010, Protección ambiental – Especies nativas de México de flora y fauna silvestres – Categorías de riesgo y especificaciones para su inclusión, exclusión o cambio – Lista de especies en riesgo. Diario Oficial de la Federación. 30 de diciembre de 2010, Segunda Sección, México.
Serna-Lagunes, R., Torres-Cantú, G. B., & García-Martínez, M. Á. (2023). Avifauna en fragmentos de bosque mesófilo de montaña y vegetación secundaria en el municipio de Huatusco, Veracruz. CienciaUAT, 1, 06–24. https://doi.org/https://doi.org/10.29059/cienciauat.v18i1.1669
Sibley, D. A. (2000). The Sibley guide to birds. New York: Alfred A. Knopf.
Skagen, S. K., Melcher, C. P., Howe, W. H., & Knopf, F. L. (1998). Comparative use of riparian corridors and oases by migrating birds in southeast Arizona. Conservation Biology, 12, 896–909. https://doi.org/https://doi.org/10.1111/j.1523-1739.1998.96384.x
Toledo-Aceves, T., García-Franco, J., Williams-Linera, G., Mac-Millan, K., & Gallardo-Hernández, C. (2014). Significance of remnant cloud forest fragments as reservoirs of tree and epiphytic bromeliad diversity. Tropical Conservation Science, 7, 230–243. https://doi.org/10.1177/194008291400700205
Toledo-Aceves, T., Meave, J., González-Espinosa, M., & Ramírez-Marcial, N. (2011). Tropical montane cloud forests: Current threats and opportunities for their conservation and sustainable management in Mexico. Journal of Environmental Management, 92, 974–981. https://doi.org/10.1016/j.jenvman.2010.11.007
Villaseñor-Gómez, J. F. (2008). Habitat use of wintering bird communities in Sonora Mexico: the importance of riparian habitats. Studies in Avian Biology, 37, 53–68.
Watson, J. E. M., Whittaker, R. J., & Dawson, T. P. (2004). Habitat structure and proximity to forest edge affect the abundance and distribution of forest-dependent birds in tropical coastal forests of southeastern Madagascar. Biological Conservation, 120, 311–327. https://doi.org/10.1016/j.biocon.2004.03.004
Williams-Linera, G., Manson, R. H., & Isunza, V. E. (2002). La fragmentación del bosque mesófilo de montaña y patrones de uso del suelo en la región oeste de Xalapa, Veracruz, México. Madera y Bosques, 8, 69–85. https://doi.org/https://doi.org/10.21829/myb.2002.811307
Wilson, S., Mitchell, G. W., Pasher, J., McGovern, M., Hudson, M. A. R., & Fahrig, L. (2017). Influence of crop type, heterogeneity and woody structure on avian biodiversity in agricultural landscapes. Ecological Indicators, 83, 218–226. https://doi.org/10.1016/j.ecolind.2017.07.059
Zarazúa-Carbajal, M., Avila-Cabadilla, L. D., Álvarez-Añorve, M. Y., Benítez-Malvido, J., & Stoner, K. E. (2017). Importance of riparian habitat for frugivorous bats in a tropical dry forest in western Mexico. Journal of Tropical Ecology, 33, 74–82. https://doi.org/10.1017/S0266467416000572

Diversidad funcional y composición de comunidades de insectos en niveles diferentes de perturbación
Functional diversity and composition of insect communities at different levels of disturbance
Víctor Manuel Caballero-Chan, Alejandra González-Moreno*, Horacio Salomón Ballina-Gómez y Carlos Juan Alvarado-López
Tecnológico Nacional de México, Instituto Tecnológico de Conkal, División de Estudios de Posgrado e Investigación, Av. Tecnológico s/n, 97345 Conkal, Yucatán, México
*Autor para correspondencia: alejandra.gonzalez@itconkal.edu.mx (A. González-Moreno)
Recibido: 18 enero 2025; aceptado: 30 mayo 2025
Resumen
La diminución en la cobertura vegetal y la perturbación antropogénica tienen efectos negativos sobre la diversidad de insectos. Este trabajo tuvo como objetivo evaluar la diversidad funcional y la composición de comunidades de insectos fitófagos y benéficos en diferentes niveles de perturbación de Yucatán, México. Se instalaron 6 trampas Malaise por sitio, durante 5 meses en temporada de lluvias. Los ejemplares se identificaron a nivel familia y grupo funcional; se analizó la diversidad en términos de riqueza, familias comunes y dominantes de cada grupo funcional. Se registraron 25,872 individuos de 106 familias, 12 órdenes y 4 grupos funcionales (fitófagos, polinizadores, depredadores y parasitoides). Aunque la riqueza de familias fue similar, la diversidad de familias comunes y dominantes mostró diferencias en los sitios con niveles medios y altos de perturbación. Estos resultados sugieren que algunas familias son exitosas en niveles altos de perturbación y otras disminuyen su diversidad. Las familias dominantes de los fitófagos, polinizadores, parasitoides y depredadores fueron: Pyralidae, Geometridae, Tachinidae y Coccinellidae, respectivamente.
Palabras clave: Urbanización; Grupos funcionales; Fitófagos; Insectos benéficos
Abstract
Decreased vegetation cover and anthropogenic disturbance can negatively impact insect diversity. This research aimed to assess the functional diversity and community composition of phytophagous and beneficial insects at different levels of disturbance in Yucatán, Mexico. Six Malaise traps were deployed at each site during 5 months in the rainy season. Specimens were identified to the family and functional group levels, and diversity was analyzed based on family richness, as well as the composition of common and dominant families for every functional group. A total of 25,872 individuals representing 106 families, 12 orders, and 4 functional groups were recorded (phytophagous, pollinators, predators and parasitoids). While family richness was comparable across sites, the diversity of common and dominant families differed between areas with medium and high levels of disturbance. These results suggest that some families thrive under high disturbance levels, whereas other experiences a decline in diversity. The dominant families of phytophagous, pollinators, parasitoids and predators were: Pyralidae, Geometridae, Tachinidae, and Coccinellidae, respectively.
Keywords: Urbanization; Functional groups; Phytophagous; Beneficial insects
Introducción
En México, un país considerado megadiverso, el crecimiento de las zonas urbanas y la intensificación de la agricultura, están poniendo en riesgo la diversidad de insectos nativos presentes en zonas naturales (Martínez-Ramos et al., 2016), así como ocasionando cambios en el comportamiento de distintos grupos de insectos, que comprometen funciones ecológicas críticas como la polinización, el control biológico de plagas y la descomposición de la materia orgánica (Wagner et al., 2021). La perturbación antropogénica derivada de la urbanización, que incluye la construcción de infraestructura y el desarrollo de asentamientos humanos, junto con la agricultura intensiva, la deforestación, la fragmentación del hábitat, la contaminación y cambio climático, está generando una presión sin precedentes sobre la biodiversidad (Betts et al., 2019; Fahrig et al., 2019). La disminución de especies de vertebrados, como aves, anfibios y mamíferos está bien documentada; sin embargo, la pérdida de diversidad en invertebrados es menos conocida y se desconoce si está ocurriendo a la misma velocidad que en otros grupos; aunque estudios más recientes, incluidos varios metaanálisis, han evidenciado el declive de los insectos (Wagner et al., 2021).
En particular, se ha demostrado que la urbanización y agricultura intensiva afectan negativamente a los polinizadores, principalmente por la pérdida de hábitat, cambios en la disponibilidad de alimentos y la alteración de sus patrones de comportamiento (Biella et al., 2022; Fisogni et al., 2020; Tavares-Brancher et al., 2024). Pero, la urbanización no solo afecta a los polinizadores, sino que en general contribuye a la disminución de insectos debido a múltiples factores, como el incremento en la intensidad lumínica (Boyes et al., 2021), en la temperatura, evaporación y la exposición al viento y a diversos contaminantes, que alteran su comportamiento y actividad (Dirzo, 2014; Wagner et al., 2021); especialmente aquellos que ocupan altos niveles tróficos como los parasitoides y depredadores (Betancourt et al., 2021; Janzen y Hallwachs, 2021). Esta pérdida de diversidad no solo es preocupante por la pérdida de especies en sí misma, sino también por la disminución de las múltiples funciones ecológicas que realizan los insectos (Fenoglio et al., 2020; Wagner et al., 2021).
En México se han realizado algunos estudios sobre cómo la diversidad de insectos varía en varios niveles de perturbación, con resultados contrastantes, dependiendo del taxón a estudiar, del grupo funcional al que pertenecen y de las características de los sitios perturbados. En general, se ha demostrado que la urbanización tiene un impacto negativo sobre la diversidad de abejas (Muñoz-Urías et al., 2025), coleópteros (Cortés-Arzola y León-Cortés, 2021) y hormigas, principalmente de hábitos arborícolas (Roche-Ortega y Castaño- Meneses, 2015) y mariposas (Ramírez-Restrepo y Halffter, 2013). Actualmente, la península de Yucatán está siendo amenazada por el crecimiento urbano descontrolado y las prácticas agrícolas no sostenibles, por lo que se planteó la siguiente pregunta de investigación ¿cómo varían las comunidades de insectos en términos de diversidad funcional y composición, en diferentes niveles de perturbación? Tomando en cuenta que los resultados de las evaluaciones de diversidad pueden variar según el nivel taxonómico considerado y los requerimientos ecológicos del taxón a estudiar (Fenoglio et al., 2020), en este trabajo se planteó abordarlo a nivel de familia, considerando el grupo funcional que conforman y la función que llevan a cabo como fitófagos, depredadores, parasitoides o polinizadores.
El objetivo del presente trabajo fue evaluar la diversidad funcional y la composición de comunidades de insectos fitófagos y benéficos en diferentes niveles de perturbación de Yucatán; para ello se plantearon las siguientes hipótesis: la diversidad de insectos fitófagos será mayor en zonas con mayor nivel de perturbación; por el contrario, los insectos benéficos serán más diversos en niveles de perturbación menores y la familia dominante de cada grupo funcional será diferente en cada nivel de perturbación. Realizar estas evaluaciones podría ser significativo para implementar estrategias de conservación dentro de las ciudades, además de que las evaluaciones de la diversidad de invertebrados son prioritarias para avanzar en el entendimiento de la defaunación, principalmente en regiones tropicales (Dirzo, 2014; Wagner et al., 2021).
Materiales y métodos
El estudio se llevó a cabo en 3 zonas de Yucatán que se eligieron considerando el grado de perturbación, de acuerdo con el índice de disturbio (ID), según el método propuesto por Martorell y Peters (2005). En cada zona, se seleccionaron 2 sitios al azar, se utilizaron fotografías aéreas (INEGI SINFA 1:250 000, 2019 CLAVE F16-10 LÍNEA 166) que fueron importadas al programa Arc view 3.1. El ID se basa en la cuantificación de 15 parámetros, los cuales están comprendidos en 1 de 3 categorías: 1) cría de ganado (frecuencia de excrementos de cabra, frecuencia de excremento de vaca, ramoneo, caminos para el ganado y compactación del suelo); 2) actividades humanas (extracción de leña, número de caminos, superficie de senderos, proximidad de asentamientos humanos, cercanía a núcleos de actividad humana, porcentaje de uso del suelo y evidencia de incendios forestales) y 3) degradación del suelo (porcentaje de erosión, presencia de islas de erosión y superficie totalmente modificada). Una vez que se estimó el índice de perturbación en cada sitio, se establecieron 3 niveles de perturbación: alto (índice de perturbación entre 10 y 15), medio (entre 2.1 y 5) y bajo (entre 0 y 2). Así, se seleccionaron 2 sitios por nivel de afectación, resultando 6 sitios en total (tabla 1).
La zona con alto nivel de perturbación, a la cual se denominó zona urbana, se ubicó alrededores de la ciudad de Mérida (21°01’ N, 89°33’ O), al norte del estado, dominada por un paisaje urbano, con 75% de cobertura gris y apenas 10% de cobertura de material vegetal (Biles y Lemberg, 2023). La zona con nivel medio de perturbación, nombrada zona periurbana, se localiza en el municipio de Conkal (21°05’ N, 89°30’ O), al este del municipio de Mérida, con 40% de cobertura gris y 35% de cobertura vegetal, dominada por cultivos de maíz y vegetación circundante de selva baja caducifolia (Rzedowski, 1978). La zona, con bajo nivel de perturbación, llamada zona natural, se localiza en la reserva privada “Komchén de los Pájaros” (21°13’ N, 89°19’ O), al oeste de los municipios de Dzemul y Telchac, presenta una cobertura vegetal de 80% y una cobertura gris inferior a 5% (fig. 1); la vegetación es selva baja caducifolia, conformada principalmente por varias especies de la familia Fabaceae: Piscidia piscipula (jabín), Caesalpinia gaumeri (kitinché), Lysiloma latisiliquum (tzalam) y la familia Burseraceae: Bursera simaruba (palo mulato) (Flores y Espejel, 1994). Las 3 zonas presentan un clima cálido subhúmedo con lluvias en verano con rangos de precipitación anual que van de 1,050 mm a 1,200 mm. La temperatura media anual es de 26 a 28 °C.
Tabla 1
Índice y categoría de perturbación de los 6 sitios seleccionados en México.
| Nombre del sitio | Tipo de sitio | Índice | Nivel |
| Mérida 1 | Huerto en ciudad | 10.33 | Alto |
| Mérida 2 | Huerto en ciudad | 11.35 | Alto |
| Conkal 1 | Huerto periurbano con cultivo de maíz | 2.22 | Medio |
| Conkal 2 | Huerto periurbano con cultivo de maíz | 3.59 | Medio |
| Komchén de los Pájaros 1 | Selva baja caducifolia | 0.78 | Bajo |
| Komchén de los Pájaros 2 | Selva baja caducifolia | 0.81 | Bajo |
Se llevaron a cabo muestreos sistemáticos durante la temporada de lluvias, de julio a diciembre de 2023. En cada nivel de perturbación se instalaron 2 trampas de intercepción tipo Malaise, separadas por más de 2 km, para asegurar la independencia de las muestras; estas trampas fueron seleccionadas por su alta eficacia en la captura pasiva de insectos voladores, incluidos lepidópteros diurnos y nocturnos (Schmidt et al., 2019), además de ser uno de los métodos de muestreo mayormente utilizados para hacer evaluaciones de biodiversidad, y permiten obtener un muestreo representativo de los ensambles presentes en cada sitio (Chan-Canché et al., 2020; Kaczmarek et al., 2022). En los sitios urbanos las trampas fueron colocadas en jardines de casas particulares; en la zona periurbana se colocaron en cultivos de maíz y en la zona natural en parches de vegetación. Las trampas funcionaron ininterrumpidamente durante 5 meses con cortes quincenales de recolecta, resultando un total de 10 muestras por sitio. Los botes recolectores de las trampas tenían 1 L de etanol desnaturalizado al 70%, el cual era reemplazado en cada recolecta. Cada muestra se procesó según las técnicas curatoriales convencionales, en el laboratorio de plagas agrícolas del Instituto Tecnológico de Conkal; todos los insectos se conservaron en etanol al 70% y se seleccionaron algunos ejemplares para su montaje en seco en alfileres entomológicos para su posterior identificación; en el caso de los lepidópteros, se empleó la técnica de relajación en cámara húmeda, extensión de alas sobre planchas entomológicas y posterior secado para su correcta preservación y manejo en la colección. Posteriormente se realizó la identificación taxonómica de los ejemplares a categoría de familia, utilizando claves especializadas en insectos de Latinoamérica, como las de Goulet y Huber (1993), Borror y White (1998), Arnett (2000), Triplehorn y Johnson (2005).
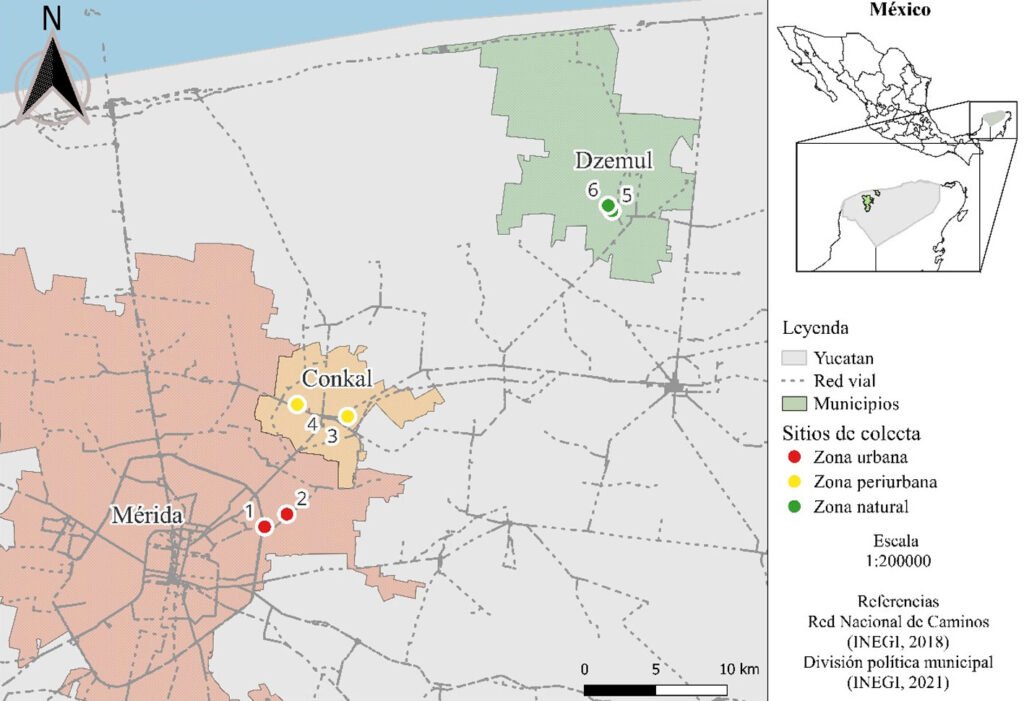
Se analizó la representatividad del muestreo con el software EstimateS 9.10 mediante curvas de acumulación, con el estimador no paramétrico jackknife 1, conocido por ser uno de los estimadores menos sesgados para muestras pequeñas (Magurran, 2004); se utilizaron las 10 fechas de recolecta como medida del esfuerzo de muestreo, con un total de 3,600 horas de recolecta por trampa. Asimismo, se analizaron las diferencias entre la riqueza de familias entre sitios, considerando los intervalos de confianza al 95% de 1,000 remuestreos calculados mediante la prueba de bootstrap (Colwell y Elsensohn, 2014); la no superposición de los intervalos de confianza indica diferencias estadísticamente significativas (Colwell, 2006). Para realizar el análisis de diversidad de insectos en cada nivel de perturbación, las familias identificadas se agruparon considerando el grupo funcional que conforman: fitófagos, polinizadores, parasitoides o depredadores; para cada grupo el análisis de diversidad fue calculado mediante medidas de diversidad verdadera, usando el software SPADE (Chao y Shen, 2010). Estas medidas contemplan 3 niveles de diversidad basadas en los números de Hill, qD (Jost, 2006): 0D, se refiere a la riqueza de familias solamente; 1D, es la diversidad ecológica si todas las familias tuvieran la misma importancia relativa, usa el inverso del exponencial de la entropía de Shannon; y 2D que considera solo a las familias dominantes, mediante el inverso del índice de Simpson (Moreno et al., 2011). Todos los valores de qD se calcularon por separado y se tomó en cuenta cada zona de manera individual (zona urbana, zona periurbana y zona natural), lo que permitió evaluar la diversidad por cada zona de perturbación para después comparar entre sitios, usando intervalos de confianza de 95% que fueron calculados mediante la prueba de bootstrap, para saber si existen diferencias significativas entre zonas, la no superposición de los intervalos de confianza indica diferencias estadísticamente significativas (Colwell, 2006).
Posteriormente, se analizó la composición de los ensambles de cada grupo funcional en términos de la distribución de abundancias de cada familia, para lo que se construyeron curvas de rango-abundancia para cada nivel de perturbación (Whittaker, 1972).
Resultados
Se recolectaron 25,872 individuos pertenecientes a 4 grupos funcionales: fitófagos, polinizadores, parasitoides y depredadores, clasificados en 12 órdenes y 106 familias (tabla 2). Las curvas de acumulación indican que el muestreo tuvo una eficiencia de 83.4%, (familias observadas = 86; jackknife 1 = 103.1) para la zona urbana, de 84.7% para la zona periurbana (familias observadas = 90; jackknife 1 = 106.2) y de 83.5 % para la zona natural (familias observadas = 73; jackknife 1 = 87.4), con menor riqueza para la zona natural, con intervalos de confianza al 95% (fig. 2).
En el grupo de los fitófagos, la riqueza de familias (0D) y la diversidad de familias dominantes (2D) no mostraron diferencias significativas entre zonas, considerando el sobrelapamiento de los intervalos de confianza al 95%; únicamente la diversidad de familias comunes (1D) muestra que la zona periurbana tiene el valor más alto (tabla 3). La familia Pyralidae se registró dominando en las zonas periurbana y natural, en cambio, la familia Cicadellidae, lo hizo únicamente en la zona urbana (fig. 3A).
Los insectos polinizadores, al igual que los fitófagos, presentaron diferencias en la diversidad de familias comunes (1D), siendo los más diversos en la zona urbana con los intervalos de confianza al 95%. La riqueza (0D) y diversidad de familias dominantes (2D) fueron similares en los 3 sitios (tabla 3). Sin embargo, pese a que la diversidad fue similar, las familias dominantes fueron diferentes en cada nivel de perturbación: Geometridae en los sitios periurbanos con maíz, Nymphalidae en el sitio natural y Erebidae en la zona urbana (fig. 3B).
Tabla 2
Grupos funcionales, órdenes y familias de insectos identificados en un gradiente de perturbación de Yucatán, México.
| Grupo | Orden | Familia | Zona urbana | Zona periurbana | Zona natural | Total |
| Fitófago | Lepidoptera | Pyralidae | 2,298 | 2,117 | 1,459 | 5,874 |
| Crambidae | 219 | 969 | 505 | 1,693 | ||
| Tortricidae | 242 | 363 | 164 | 769 | ||
| Noctuidae | 3 | 286 | 23 | 312 | ||
| Tineidae | 12 | 116 | 54 | 182 | ||
| Oecophoridae | 0 | 1 | 0 | 1 | ||
| Hemiptera | Cicadellidae | 3,063 | 1,034 | 281 | 4,378 | |
| Diaspididae | 309 | 24 | 4 | 337 | ||
| Psyllidae | 1 | 122 | 2 | 125 | ||
| Aphididae | 43 | 11 | 19 | 73 | ||
| Derbidae | 9 | 25 | 3 | 37 | ||
| Cicadidae | 13 | 8 | 10 | 31 | ||
| Cixiidae | 9 | 7 | 7 | 23 | ||
| Liviidae | 19 | 0 | 1 | 20 | ||
| Rhyparochromidae | 1 | 18 | 0 | 19 | ||
| Berytidae | 7 | 3 | 0 | 10 | ||
| Delphacidae | 2 | 6 | 1 | 9 | ||
| Alydidae | 3 | 4 | 0 | 7 | ||
| Membracidae | 3 | 1 | 1 | 5 | ||
| Cercopidae | 4 | 0 | 0 | 4 | ||
| Coreidae | 0 | 4 | 0 | 4 | ||
| Miridae | 0 | 2 | 1 | 3 | ||
| Pentatomidae | 1 | 2 | 0 | 3 | ||
| Pyrrhocoridae | 0 | 3 | 0 | 3 | ||
| Lygaeidae | 0 | 2 | 0 | 2 | ||
| Triozidae | 0 | 0 | 2 | 2 |
| Tropiduchidae | 1 | 1 | 0 | 2 | ||
| Cydnidae | 1 | 0 | 0 | 1 | ||
| Dictyopharidae | 0 | 1 | 0 | 1 | ||
| Rhopalidae | 0 | 1 | 0 | 1 | ||
| Tingidae | 1 | 0 | 0 | 1 | ||
| Diptera | Drosophilidae | 496 | 284 | 98 | 878 | |
| Ulidiidae | 158 | 71 | 47 | 276 | ||
| Tephritidae | 1 | 11 | 8 | 20 | ||
| Psilidae | 1 | 0 | 0 | 1 | ||
| Coleoptera | Chrysomelidae | 48 | 62 | 34 | 144 | |
| Curculionidae | 20 | 19 | 14 | 53 | ||
| Mordellidae | 9 | 1 | 3 | 13 | ||
| Bruchidae | 0 | 2 | 0 | 2 | ||
| Orthoptera | Gryllidae | 5 | 15 | 57 | 77 | |
| Acrididae | 1 | 22 | 6 | 29 | ||
| Tettigoniidae | 1 | 6 | 2 | 9 | ||
| Thysanoptera | Phlaeothripidae | 2 | 13 | 0 | 15 | |
| Thripidae | 1 | 3 | 3 | 7 | ||
| Polinizador | Lepidoptera | Geometridae | 187 | 1,672 | 222 | 2,081 |
| Erebidae | 251 | 1,214 | 390 | 1,855 | ||
| Nymphalidae | 25 | 257 | 1,127 | 1,409 | ||
| Pieridae | 77 | 122 | 119 | 318 | ||
| Hesperiidae | 31 | 82 | 58 | 171 | ||
| Pterophoridae | 12 | 38 | 64 | 114 | ||
| Lycaenidae | 23 | 66 | 14 | 103 | ||
| Riodinidae | 2 | 61 | 0 | 63 | ||
| Sphingidae | 14 | 8 | 1 | 23 | ||
| Diptera | Bombyliidae | 25 | 459 | 66 | 550 | |
| Syrphidae | 25 | 103 | 7 | 135 | ||
| Tipulidae | 6 | 25 | 25 | 56 | ||
| Hymenoptera | Apidae | 25 | 14 | 7 | 46 | |
| Halictidae | 1 | 0 | 0 | 1 | ||
| Parasitoide | Diptera | Tachinidae | 106 | 593 | 158 | 857 |
| Pipunculidae | 13 | 18 | 15 | 46 | ||
| Hymenoptera | Braconidae | 146 | 99 | 78 | 323 | |
| Ichneumonidae | 37 | 114 | 63 | 214 | ||
| Encyrtidae | 114 | 28 | 14 | 156 | ||
| Figitidae | 67 | 2 | 2 | 71 | ||
| Chalcididae | 21 | 23 | 3 | 47 | ||
| Bethylidae | 31 | 4 | 11 | 46 | ||
| Eulophidae | 22 | 5 | 6 | 33 | ||
| Eupelmidae | 14 | 5 | 5 | 24 | ||
| Diapriidae | 12 | 4 | 5 | 21 | ||
| Aphelinidae | 12 | 5 | 2 | 19 | ||
| Pteromalidae | 12 | 4 | 3 | 19 | ||
| Mymaridae | 3 | 0 | 11 | 14 | ||
| Eurytomidae | 9 | 3 | 0 | 12 | ||
| Evaniidae | 5 | 2 | 3 | 10 | ||
| Tiphiidae | 4 | 4 | 1 | 9 | ||
| Trichogrammatidae | 5 | 0 | 3 | 8 | ||
| Perilampidae | 4 | 1 | 1 | 6 | ||
| Chrysididae | 0 | 5 | 0 | 5 | ||
| Platygastridae | 3 | 1 | 0 | 4 | ||
| Eucharitidae | 3 | 0 | 0 | 3 | ||
| Tetracampidae | 0 | 1 | 2 | 3 | ||
| Mutillidae | 0 | 2 | 0 | 2 | ||
| Dryinidae | 0 | 0 | 1 | 1 | ||
| Rhopalosomatidae | 0 | 1 | 0 | 1 | ||
| Depredador | Coleoptera | Coccinellidae | 451 | 118 | 51 | 620 |
| Dytiscidae | 5 | 2 | 1 | 8 | ||
| Carabidae | 2 | 3 | 0 | 5 | ||
| Diptera | Dolichopodidae | 250 | 41 | 25 | 316 | |
| Asilidae | 43 | 140 | 59 | 242 | ||
| Scenopinidae | 0 | 4 | 0 | 4 | ||
| Hemiptera | Anthocoridae | 2 | 2 | 1 | 5 | |
| Reduviidae | 1 | 3 | 0 | 4 | ||
| Stenocephalidae | 1 | 0 | 0 | 1 | ||
| Hymenoptera | Crabronidae | 58 | 16 | 18 | 92 | |
| Vespidae | 20 | 22 | 4 | 46 | ||
| Pompilidae | 5 | 8 | 2 | 15 | ||
| Sphecidae | 3 | 2 | 0 | 5 | ||
| Sapygidae | 0 | 2 | 1 | 3 | ||
| Thynnidae | 0 | 2 | 0 | 2 | ||
| Scoliidae | 0 | 0 | 1 | 1 | ||
| Mantodea | Mantidae | 9 | 17 | 8 | 34 | |
| Mantoididae | 2 | 1 | 0 | 3 | ||
| Mecoptera | Bittacidae | 1 | 0 | 0 | 1 | |
| Neuroptera | Chrysopidae | 24 | 73 | 31 | 128 | |
| Berothidae | 0 | 0 | 1 | 1 | ||
| Myrmeleontidae | 0 | 0 | 1 | 1 |
Los parasitoides, mostraron mayores diferencias en términos de diversidad, siendo la zona urbana la que tuvo la mayor diversidad de especies comunes (1D) y dominantes (2D), pese que la riqueza de familias (0D) fue similar en los 3 niveles de perturbación, según los valores de los intervalos de confianza al 95% (tabla 3). La familia Tachinidae, contrario a lo esperado, se registró como dominante en la zona periurbana y natural, pero en la zona urbana, la familia Braconidae fue la dominante (fig. 3C).
Los insectos depredadores presentaron la misma riqueza de familias (0D) en los diferentes sitios, pero fueron más diversos (1D y 2D) en la zona natural y periurbana, de acuerdo con los intervalos de confianza al 95% (tabla 3) con la familia Asilidae dominando las comunidades más diversas; por el contrario, Coccinellidae dominó las comunidades de la zona urbana (fig. 3D).
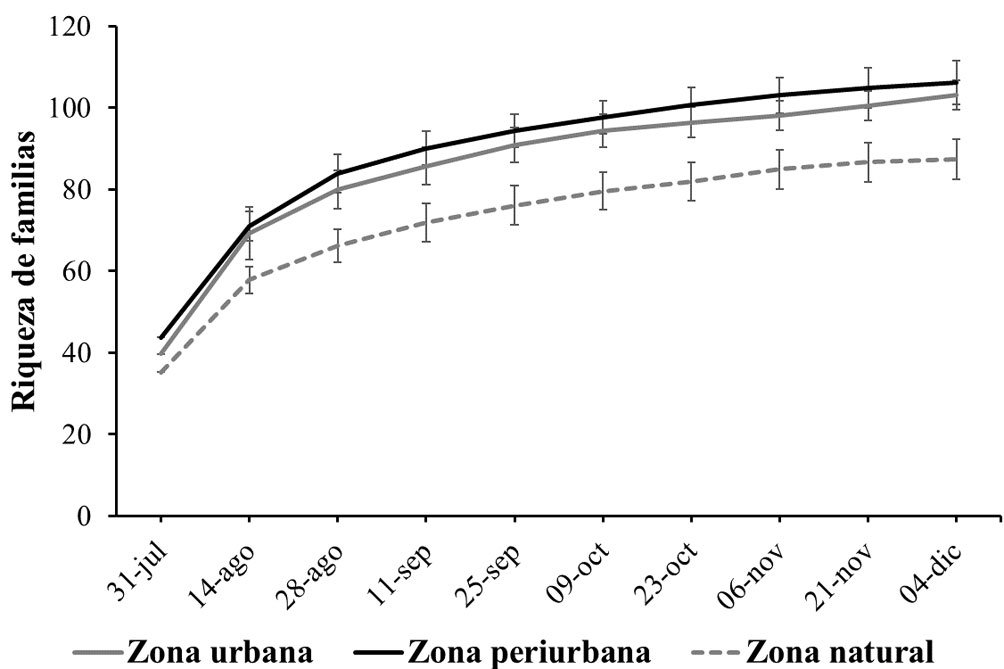
Discusión
Los resultados de este trabajo fueron contrarios a nuestra hipótesis de investigación, ya que ésta sugiere que en zonas con altos niveles de perturbación como las ciudades, habría menor diversidad de insectos, principalmente especialistas como los parasitoides, considerando la hipótesis del aumento del disturbio, que indica una disminución en la riqueza de artrópodos, particularmente especialistas, conforme aumenta el grado de urbanización (Gray, 1989, en Fenoglio et al., 2020) y la teoría sobre la complejidad estructural de la vegetación, que afirma que a mayor cobertura vegetal en ecosistemas, habrá un mayor número de plantas disponibles, lo que permitirá alojar mayor diversidad de fitófagos y, por consiguiente, de depredadores y parasitoides (González-Moreno et al., 2023; Guo et al., 2021; Neal et al., 2024).
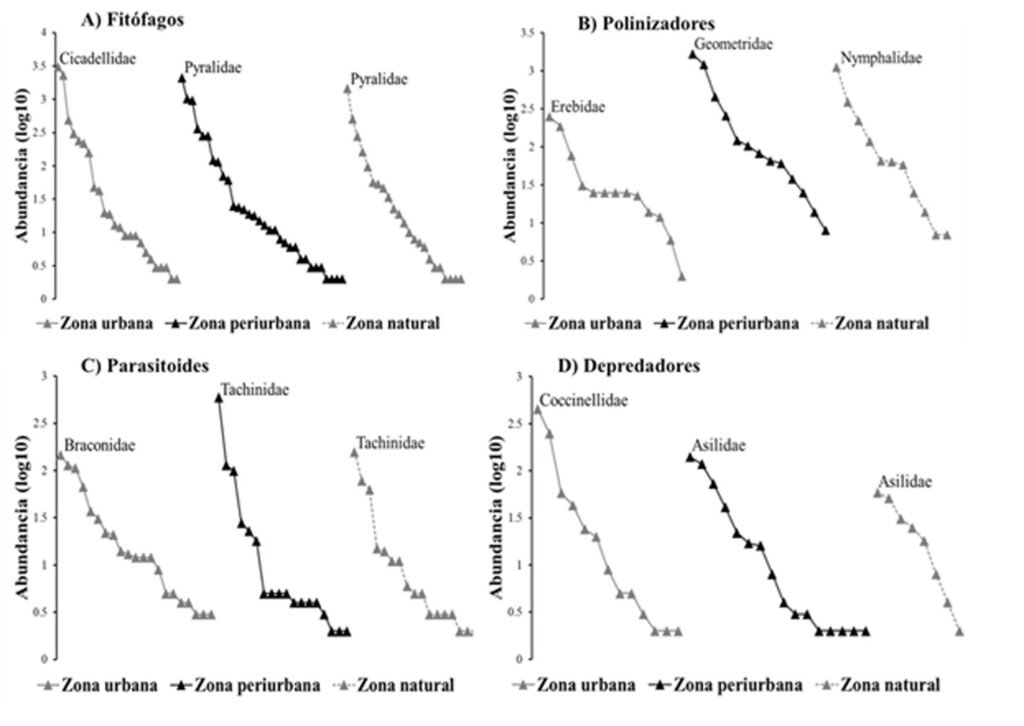
Tabla 3
Diversidad verdadera de grupos funcionales de insectos, como número efectivo de familias, con números de Hill para estimar la riqueza de familias (0D), la diversidad de familias comunes (1D) y la diversidad de familias dominantes (2D) en 3 zonas con diferente nivel de perturbación de Yucatán. *Los intervalos de confianza (IDC) al 95%, indican diferencias significativas.
| Grupos funcionales | Índices de diversidad verdadera | ||
| 0D (95% IDC) | 1D (95% IDC) | 2D (95% IDC) | |
| Fitófagos | |||
| Zona urbana | 50.6 (39.9, 84.8) | 4.8 (4.6, 4.9)* | 3.2 (2.4, 4.1) |
| Zona periurbana | 42.4 (39.0, 56.7) | 7.2 (6.9, 7.4)* | 4.7 (4.0, 5.3) |
| Zona natural | 30.3 (27.7, 43.1) | 5.3 (5.0, 5.6)* | 3.1 (2.5, 3.8) |
| Polinizadores | |||
| Zona urbana | 14.9 (14.1, 25.2) | 6.7 (6.2, 7.2)* | 4.6 (4.2, 5.0) |
| Zona periurbana | 13.0 (13.0, 13.0) | 5.2 (5.0, 5.4)* | 3.7 (3.1, 4.3) |
| Zona natural | 12.5 (12.0, 20.0) | 4.4 (4.2, 4.7)* | 2.9 (2.3, 3.6) |
| Parasitoides | |||
| Zona urbana | 21.0 (21.0, 21.0) | 10.8 (10.0, 11.6)* | 7.7 (7.4, 8.0)* |
| Zona periurbana | 23.6 (22.3, 32.0) | 4.0 (3.6, 4.3)* | 2.3 (1.5, 3.1)* |
| Zona natural | 21.3 (20.2, 29.3) | 6.9 (6.0, 7.8)* | 4.2 (3.6, 4.8)* |
| Depredadores | |||
| Zona urbana | 18.7 (16.4, 31.7) | 4.1 (3.8, 4.5)* | 2.8 (2.0, 3.6)* |
| Zona periurbana | 17.5 (17.0, 22.7) | 7.0 (6.3, 7.7) | 5.0 (4.5, 5.6) |
Las diferencias encontradas en la riqueza de familias de insectos, sin tomar en cuenta su función en los ecosistemas, puede explicarse según la hipótesis de la perturbación media, que propone que la diversidad puede ser mayor en sitios donde la perturbación no es muy frecuente ni muy intensa, comparada con sitios no perturbados o con perturbación intensa (Connell, 1978).
El efecto de la perturbación y heterogeneidad del paisaje sobre la diversidad puede ser diferente dependiendo de la escala de estudio (Corcos et al., 2019), del taxón y del grupo funcional (Fenoglio et al., 2020), como se pudo observar en este trabajo. Las comunidades más diversas de fitófagos se presentaron en los sitios periurbanos con cultivos de maíz, probablemente, por la oferta mayor de alimento que puede representar el cultivo, facilitando el acceso a recursos alimenticios y refugio, que a su vez, favorece una mayor equidad de familias comunes (Landry et al., 2020); además, si consideramos nuevamente la hipótesis del disturbio medio, los sitios periurbanos con un nivel medio de perturbación, estarían alojando mayor diversidad, en este caso de fitófagos. Asimismo, se ha demostrado que zonas perturbadas que integran espacios verdes como parques urbanos, jardines residenciales, huertos verticales y familiares, parcelas de policultivos y parches de vegetación natural, favorecen la diversidad de fitófagos, al proporcionarles recursos alimenticios y refugios suficientes, que les permite adaptarse y prosperar en hábitats alterados por la actividad humana (Landry et al., 2020; Ruiz-Montoya et al., 2014). En los ensambles, la familia Pyralidae fue dominante en las zonas periurbana y natural, debido a su capacidad para aprovechar tanto plantas cultivadas como nativas, lo que le permite alimentarse y completar su ciclo de vida con eficacia (Cepeda, 2017). En cambio, las comunidades de la zona urbana estuvieron dominadas por la familia Cicadellidae, lo que refleja su adaptación a ambientes con altos niveles de perturbación (Trivellone et al., 2021).
La mayor diversidad de polinizadores en ciudad puede estar relacionada con la variabilidad y abundancia de recursos florales de los jardines, parques y áreas verdes presentes en los sitios, al proporcionar ciertas fuentes polínicas que favorecen dicha diversidad; estos resultados son contrarios a la que esperábamos si se considera la teoría sobre el espectro de polinización, que afirma, que a mayor cobertura vegetal en los ecosistemas, habrá un mayor número de plantas con flores, lo que permitirá alojar mayor diversidad de polinizadores. Sin embargo, es importante señalar que esta diversidad estuvo representada por familias de lepidópteros y no por abejas, debido probablemente a que estas últimas son de los grupos más sensibles a la contaminación de las ciudades (Roguz et al., 2023). Aunque se ha demostrado que en áreas urbanas, cuando se crean nuevos hábitats o refugios, como hoteles para polinizadores, jardines florales y corredores de flores silvestres, la diversidad de polinizadores tiende a incrementarse (Francini et al., 2022; Persson et al., 2023); pero el grupo de polinizadores varía dependiendo de ciertos factores asociados a la urbanización, por ejemplo, los abejorros son muy sensibles a la contaminación de las ciudades (Roguz et al., 2023); pero otros grupos pueden tener la capacidad de alimentarse en entornos urbanos (McLeod et al., 2021), como las mariposas (Lepidoptera: Papilionoidea) que son más tolerantes a la urbanización que otros polinizadores como sírfidos, moscas abejorros (Diptera: Syrphidae, Bombyliidae) y abejas (Hymenoptera: Apoidea) (Ávalos-Hernández et al., 2024). Es importante señalar que la diversidad observada es de familias consideradas comunes, lo que está reflejando la adaptación de ciertos polinizadores generalistas, que se adaptan rápidamente a las condiciones altamente perturbadas y son capaces de aprovechar la oferta floral de estos sitios (Deguines et al., 2016; Neumann et al., 2024). Las familias dominantes encontradas en este trabajo se han registrado interactuando con varias especies de plantas en jardines urbanos, realizando la función de polinización (Wonderlin et al., 2019). Geometridae fue dominante en los huertos periurbanos con maíz, probablemente porque son lepidópteros que disminuyen en sitios más urbanizados (Gaona et al., 2021); Nymphalidae fue dominante en la vegetación natural y Erebidae en la ciudad; esto es relevante porque las áreas urbanas y agrícolas alteran los patrones de comportamiento de los lepidópteros nocturnos debido a la luz artificial, mientras que los lepidópteros diurnos en áreas naturales reflejan su dependencia de hábitats con bajo impacto humano (Seymoure, 2018).
La diversidad de parasitoides encontrada, también fue contraria a los patrones esperados, ya que, a mayor cobertura vegetal en los ecosistemas, habrá un mayor número de hospederos disponibles, lo que permitirá alojar mayor diversidad de parasitoides (Parsons y Frank, 2019). Esto puede explicarse por la denso-dependencia de los parasitoides a sus hospederos, incluso en ciudades (Rocha y Fellowes, 2018), probablemente, porque las trampas se colocaron en jardines de casas particulares, los cuales tenían diferentes especies vegetales, ornamentales, frutales y arbustivas que ofrecían microhábitats y recursos alimenticios a los hospederos y por consiguiente a sus parasitoides (Klaus et al., 2024; Lucatero et al., 2024; Start et al., 2020). Otra razón de los valores elevados de diversidad en la zona urbana, puede explicarse por la habilidad de dispersión de los bracónidos, ya que se ha comprobado que los artrópodos que ocupan altos niveles tróficos como depredadores y parasitoides, serán exitosos en sitios urbanos, si tienen alta capacidad de dispersión (Korányi et al., 2022). Tachinidae, que no ha sido registrada como una familia particularmente abundante para la región, fue dominante en los huertos periurbanos con maíz y la vegetación natural; además no es una familia de parasitoides hiperdiversa en comparación con los himenópteros parasitoides, por lo que es menos probable de encontrarse como dominante en los sitios (Kankonda et al., 2018). Por otra parte, se ha registrado que la abundancia de esta familia disminuye en áreas con altas densidades de edificios y calles que actúan como barreras que dificultan la dispersión de los individuos, así como la localización de sus hospederos (Corcos et al., 2019). Por el contrario, la familia Braconidae dominó los sitios de zonas urbanas, lo que podría sugerir su adaptación a ambientes altamente perturbados (Koptur et al., 2024), además de que se ha demostrado que la urbanización favorece especies generalistas capaces de explotar diferentes recursos, como podría ser el caso de Braconidae, parasitoides con mayor variedad en estrategias de desarrollo y biología.
Los depredadores fueron más diversos en áreas periurbanas con maíz y selva, al contrario de los parasitoides, que fueron más diversos en ciudad, probablemente porque al ocupar nichos similares, estén evitando la competencia. Cabe destacar que los depredadores, estuvieron representados mayoritariamente por diferentes familias del orden Coleoptera, que es uno de los grupos de insectos más afectados por la urbanización en términos de riqueza de especies, pero no de abundancia (Fenoglio et al., 2020). Aunque los coleópteros depredadores pueden aprovechar más eficientemente la oferta extendida del alimento que otros enemigos naturales, al adaptarse mejor a los cambios ambientales por urbanización (Gardiner et al., 2021; Liere y Cowal, 2024); como se observó en nuestros resultados, Coccinellidae fue dominante únicamente en la zona urbana, lo que confirma que son los depredadores mejor adaptados a la urbanización, por su capacidad para prosperar en las condiciones microclimáticas características de este ambiente (Kawakami et al., 2016; Meseguer et al., 2024). En cambio, Asilidae fue dominante en las zonas periurbana con maíz y natural, lo que puede reflejar su mayor supervivencia en zonas menos modificadas por la actividad humana (Pascacio-Villafán y Cohen, 2023).
Las diferencias de diversidad observadas para los diferentes ensambles de insectos, reflejan que las zonas urbanas pueden inducir cambios en las comunidades de insectos, e incluso, en algunos casos, favorecer esta diversidad. Pero, esto solo funcionará para cierto grupo de organismos que tengan estrategias tipo “r”, con hábitos generalistas y alta capacidad de dispersión y adaptación, que los hace exitosos en ambientes urbanos (Martinson y Raupp, 2013), como ciertas especies de avispas que pueden ser resistentes a la urbanización (Christie y Hochuli, 2009), como en nuestros resultados representadas por Braconidae; también familias que estén mejor adaptadas a ambientes con altos niveles de perturbación y que pueden colonizar nuevos hábitats creados por la actividad humana y volverse dominantes dentro de las comunidades al verse favorecidas por factores ambientales como la temperatura y la humedad (Adams et al., 2020; Sire et al., 2022). Sin embargo, los resultados contrastantes reportados en la literatura sobre los efectos de la urbanización sobre las comunidades de artrópodos (Arnold, 2022), sugieren que es prioritario continuar con esta línea de investigación.
En conclusión, la riqueza de insectos en sitios con distintos niveles de perturbación fue similar; sin embargo, se encontraron diferencias en la diversidad de familias comunes, esto resulta relevante debido a que los análisis se realizaron a un nivel taxonómico alto, de familia, donde normalmente es difícil detectar variaciones significativas en diversidad. Cabe destacar, que las diferencias fueron con las familias consideradas comunes, que probablemente sean las que presentan biologías generalistas que les permite adaptarse a condiciones adversas como las que existen en zonas perturbadas.
Contrario a lo esperado, la mayor diversidad de insectos no se encontró en la vegetación natural, sino que fue en los sitios periurbanos con cultivos de maíz, con niveles de perturbación moderados, con la familia Pyralidae dominando las comunidades. En la zona urbana, con el mayor grado de perturbación, los grupos de insectos más diversos fueron parasitoides y polinizadores con las familias Tachinidae y Erebidae, representando la dominancia de cada grupo funcional, respectivamente. Por el contrario, en la zona de menor perturbación, como fue la vegetación natural, únicamente los depredadores presentaron la mayor diversidad de insectos, con la familia Asilidae como dominante.
Agradecimientos
Los autores agradecen al Tecnológico Nacional de México por el financiamiento del proyecto “Huertos familiares y conservación de la diversidad de entomofauna benéfica” (clave: 20070.24-P) y al Consejo Nacional de Humanidades, Ciencia y Tecnología por la beca de posgrado otorgada al primer autor. También agradecemos a Xiomara Gálvez Aguilera, directora de la asociación Caribbean Conservation Coastal Ecosistem A.C., por permitirnos el acceso a la reserva privada “Komchen de los pájaros”.
Referencias
Adams, B. J., Li, E., Bahlai, C. A., Meineke, E. K., McGlynn, T. P. y Brown, B. V. (2020). Local and landscape scale variables shape insect diversity in an urban biodiversity hotspot. Ecological Applications, 30, e02089. https://doi.org/10.1002/eap.2089
Ávalos-Hernández, O., Trujano-Ortega, M., Ortega-Álvarez, R., Martínez-Fuentes, R. G., Calderón-Parra, R., García-Luna, F. et al. (2024). How does urbanization affect the fauna of the largest urban forest in Mexico? Urban Forestry and Urban Greening, 92, 128191. https://doi.org/10.1016/j.ufug.2023.128191
Arnold, J.E. (2022). Biological control services from parasitic Hymenoptera in urban agriculture. Insects, 13, 467. https://doi.org/10.3390/insects13050467
Arnett Jr., R. H. (2000). American insects: a handbook of the insects of America North of Mexico. Boca Ratón, Florida: CRC Press. https://doi.org/10.1201/9781482273892
Betancourt, E. O. M., Batis, B. V., Quiala, A. P., García, Y. M. R., Madariaga, M. C. y González, R. M. (2021). Diversidad de insectos benéficos asociada a la flora existente en fincas suburbanas en Santiago de Cuba, Cuba. Revista Chilena de Entomología, 47, 121–145. https://doi.org/10.35249/rche.47.1.21.13
Betts, M. G., Wolf, C., Pfeifer, M., Banks-Leite, C., Arroyo-Rodríguez, V., Ribeiro, D. B. et al. (2019). Extinction filters mediate the global effects of habitat fragmentation on animals. Science, 366, 1236–1239. https://doi.org/10.1126/science.aax9387
Biella, P., Tommasi, N., Guzzetti, L., Pioltelli, E., Labra, M. y Galimberti, A. (2022). City climate and landscape structure shape pollinators, nectar and transported pollen along a gradient of urbanization. Journal of Applied Ecology, 59, 1586–1595. https://doi.org/10.1111/1365-2664.14168
Biles, J. J. y Lemberg, D. S. (2023). A multi-scale analysis of urban warming in residential areas of a Latin American city: the case of Mérida, Mexico. Journal of Planning Education and Research, 43, 881–896. https://doi.org/10.1177/0739456X20923002
Borror, D. J. y White, R. E. (1998). A field guide to insects: America North of Mexico (Peterson field guides). Boston: Houghton Mifflin Harcourt.
Boyes, D. H., Evans, D. M., Fox, R., Parsons, M. S. y Pocock, M. J. (2021). Is light pollution driving moth population declines? A review of causal mechanisms across the life cycle. Insect Conservation and Diversity, 14, 167–187. https://doi.org/10.1111/icad.12447
Cepeda, D. E. (2017). Introducción a los Phycitinae de Chile (Lepidoptera: Pyralidae), nuevo registro y descripción de una nueva especie del género Homoeographa Ragonot, 1888. Insecta Mundi, 556, 1–9.
Chan-Canché, R., Ballina-Gómez, H., Leirana-Alcocer, J., Bordera, S. y González-Moreno, A. (2020). Sampling of parasitoid Hymenoptera: influence of the height on the ground. Journal of Hymenoptera Research, 78, 19–31. https://doi.org/10.3897/jhr.78.54309
Chao, A. y Shen, T.J. (2010). User’s guide for program SPADE (Species Prediction and Diversity Estimation). Taiwan: National Tsing Hua University.
Christie, F. J. y Hochuli, D. F. (2009). Responses of wasp communities to urbanization: effects on community resilience and species diversity. Journal of Insect Conservation, 13, 213–221. https://doi.org/10.1007/s10841-008-9146-5
Colwell, R. K. (2006). EstimateS: statistical estimation of species richness and shared species from samples. Versión 8 [consultado 18 Abr 2025]. Recuperado de: http://purl.oclc.org/estimates
Colwell, R. K. y Elsensohn, J. E. (2014). EstimateS turns 20: Statistical estimation of species richness and shared species from samples, with non parametric extrapolation. Ecography, 37, 609–613.
Connell, J. (1978). Diversity in tropical rain forests and coral reefs. Science, 199, 1304–1310. https://doi.org/10.1126/science.199.4335.1302
Corcos, D., Cerretti, P., Caruso, V., Mei, M., Falco, M. y Marini, L. (2019). Impact of urbanization on predator and parasitoid insects at multiple spatial scales. Plos One, 14, e0214068. https://doi.org/10.1371/journal.pone.0214068
Cortés-Arzola, S. V. y León-Cortés, J. L. (2021). Response of beetle assemblages (Insecta: Coleoptera) to patch characteristics and habitat complexity in an ever-expanding urban landscape in the Yucatán Peninsula, Mexico. Annals of the Entomological Society of America, 114, 511–521. https://doi.org/10.1093/aesa/saab017
Deguines, N., Julliard, R., De Flores, M. y Fontaine, C. (2016). Functional homogenization of flower visitor communities with urbanization. Ecology and Evolution, 6, 1967–1976. https://doi.org/10.1002/ece3.2009
Dirzo, R., Young, H. S., Galetti, M., Ceballos, G., Isaac, N. J. y Collen, B. (2014). Defaunation in the Anthropocene. Science, 345, 401–406. https://doi.org/10.1126/science.1251817
Fahrig, L., Arroyo-Rodríguez, V., Bennett, J. R., Boucher-Lalonde, V., Cazetta, E., Currie, D. J. et al. (2019). Is habitat fragmentation bad for biodiversity? Biological Conservation, 230, 179–186. https://doi.org/10.1016/j.biocon.2018.12.026
Fenoglio, M. S., Rossetti, M. R. y Videla, M. (2020). Negative effects of urbanization on terrestrial arthropod communities: a meta-analysis. Global Ecology and Biogeography, 29, 1412–1429. https://doi.org/10.1111/geb.13107
Fisogni, A., Hautekèete, N., Piquot, Y., Brun, M., Vanappelghem, C., Michez, D. et al. (2020). Urbanization drives an early spring for plants but not for pollinators. Oikos, 129, 1681–1691. https://doi.org/10.1111/oik.07274
Flores, J. S. y Espejel, I. (1994). Tipos de vegetación de la Península de Yucatán. Etnoflora yucatanense, Fascículo 3. Mérida: Universidad Autónoma de Yucatán.
Francini, A., Romano, D., Toscano, S. y Ferrante, A. (2022). The contribution of ornamental plants to urban ecosystem services. Earth, 3, 1258–1274. https://doi.org/10.3390/earth
3040071
Gaona, F. P., Íñiguez-Armijos, C., Brehm, G., Fiedler, K. y Espinosa, C. I. (2021). Drastic loss of insects (Lepidoptera: Geometridae) in urban landscapes in a tropical biodiversity hotspot. Journal of Insect Conservation, 25, 395–405. https://doi.org/10.1007/s10841-021-00308-9
Gardiner, M. M., Perry, K. I., Riley, C. B., Turo, K. J., Delgado-de la Flor, Y. A. y Sivakoff, F. S. (2021). Community science data suggests that urbanization and forest habitat loss threaten aphidophagous native lady beetles. Ecology and Evolution, 11, 2761–2774. https://doi.org/10.1002/ece3.7229
González-Moreno, A., Bordera, S., Ballina-Gómez, H. y Leirana-Alcocer, J. (2023). Age matters: variations in parasitoid diversity along a successional gradient in a dry semi-deciduous tropical forest. Bulletin of Entomological Research, 113, 604–614. https://doi.org/10.1017/S0007485323000287
Goulet, H. y Huber, J. T. (1993). Hymenoptera of the World: an identification guide to families. Ottawa: Agriculture Canada, Research Branch.
Guo, P. F., Wang, M. Q., Orr, M., Li, Y., Chen, J.T., Zhou, Q. S. et al. (2021). Tree diversity promotes predatory wasps and parasitoids but not pollinator bees in a subtropical experimental forest. Basic and Applied Ecology, 53, 134–142. https://doi.org/10.1016/j.baae.2021.03.007
Janzen, D. H. y Hallwachs, W. (2021). To us insectometers, it is clear that insect decline in our Costa Rican tropics is real, so let’s be kind to the survivors. Proceedings of the National Academy of Sciences, 118, e2002546117. https://doi.org/10.1073/pnas.2002546117
Jost, L. (2006). Entropy and diversity. Oikos, 113, 363–375. https://doi.org/10.1111/j.2006.0030-1299.14714.x
Kaczmarek, M., Entling, M. H. y Hoffmann, C. (2022). Using malaise traps and metabarcoding for biodiversity assessment in vineyards: Effects of weather and trapping effort. Insects, 13, 507. https://doi.org/10.3390/insects13060507
Kankonda, O. M., Akaibe, B. D., Sylvain, N. M. y Le Ru, B. P. (2018). Response of maize stemborers and associated parasitoids to the spread of grasses in the rainforest zone of Kisangani, DR Congo: effect on stemborers biological control. Agricultural and Forest Entomology, 20, 150–161. https://doi.org/10.1111/afe.12238
Kawakami, Y., Yamazaki, K. y Ohashi, K. (2016). Population dynamics, seasonality and aphid prey of Cheilomenes sexmaculata (Coleoptera: Coccinellidae) in an urban park in central Japan. European Journal of Entomology, 113, 192–199. https://doi.org/10.14411/eje.2016.023
Klaus, F., Tscharntke, T. y Grass, I. (2024). Trophic level and specialization moderate effects of habitat loss and land-
scape diversity on cavity nesting bees, wasps, and their parasitoids. Insect Conservation and Diversity, 17, 65–76. https://doi.org/10.1111/icad.12688
Koptur, S., Primoli, A. S., Paulino-Neto, H. F. y Whitfield, J. (2024). Pierid butterflies, legume hostplants, and parasitoids in urban areas of Southern Florida. Insects, 15, 123. https://doi.org/10.3390/insects15020123
Korányi, D., Egerer, M., Rusch, A., Szabó, B. y Batáry, P. (2022). Urbanization hampers biological control of insect pests: a global meta-analysis. Science of the Total Environment, 834, 155396. https://doi.org/10.1016/j.scitotenv.2022.155396
Landry, B., Basset, Y., Hebert, P. D. y Maes, J. M. (2020). On the Pyraloidea fauna of Nicaragua. Tropical Lepidoptera Research, 30, 93–102.
Liere, H. y Cowal, S. (2024). Local and landscape factors differentially influence predatory arthropods in urban agroecosystems. Ecosphere, 15, e4816. https://doi.org/10.
1002/ecs2.4816
Lucatero, A., Smith, N. R., Bichier, P., Liere, H. y Philpott, S. M. (2024). Shifts in host–parasitoid networks across community garden management and urban landscape gradients. Ecosphere, 15, e4833. https://doi.org/10.1002/ecs2.4833
Magurran, A. E. (2004). Measuring biological diversity. Oxford: Blackwell Publishing.
Martínez-Ramos, M., Pingarroni, A., Rodríguez-Velázquez, J., Toledo-Chelala, L., Zermeño-Hernández, I. y Bongers, F. (2016). Natural forest regeneration and ecological restoration in human modified tropical landscapes. Biotropica, 48, 745–757. https://doi.org/10.1111/btp.12382
Martinson, H. M. y Raupp, M. J. (2013). A meta-analysis of the effects of urbanisation on ground beetle communities. Ecosphere, 4, 1–24. https://doi.org/10.1890/ES12-00262.1
Martorell, C. y Peters, E. M. (2005). The measurement of chronic disturbance and its effects on the threatened cactus Mammillaria pectinifera. Biological Conservation, 124, 199–207. https://doi.org/10.1016/j.biocon.2005.01.025
Mc Leod, B. C., Águila, M. K., Zegers, M. G. y Cárcamo, G. J. (2021). Refugio u “Hoteles de Insectos”, simulación de hábitat para el establecimiento de fauna auxiliar. Punta Arenas, Chile: Informativo INIA Kampenaike. Núm. 110. Recuperado el 15 de enero, 2025 de: https://hdl.handle.net/20.500.14001/67359
Meseguer, R., Madeira, F., Kavallieratos, N. G. y Pons, X. (2024). Phenology, population trends and natural enemy complex of Illinoia liriodendri in Spain. Phytoparasitica, 52, 40. https://doi.org/10.1007/s12600-024-01145-7
Moreno, C. E., Barragán, F., Pineda, E. y Pavón, N. P. (2011). Reanálisis de la diversidad alfa: alternativas para interpretar y comparar información sobre comunidades ecológicas. Revista Mexicana de Biodiversidad, 82, 1249–1261. https://doi.org/10.22201/ib.20078706e.2011.4.745
Muñoz-Urias, A., Araujo-Alanis, L., Huerta-Martínez, F. M., Jacobo-Pereira, C. y Razo-León, A. E. (2025). Effects of urbanization and floral diversity on the bee community (Hymenoptera, Apoidea) in an oak forest in a Protected Natural Area of Mexico. Journal of Hymenoptera Research, 98, 47–68. https://doi.org/10.3897/jhr.98.131191
Neal, W., Araya, Y. y Wheeler, P. M. (2024). Influence of canopy structural complexity on urban woodland butterfly species richness. Journal of Insect Conservation, 28, 1051–1062. https://doi.org/10.1007/s10841-024-00594-z
Neumann, A. E., Conitz, F., Karlebowski, S., Sturm, U., Schmack, J. M. y Egerer, M. (2024). Flower richness is key to pollinator abundance: the role of garden features in cities. Basic and Applied Ecology, 79, 102–113. https://doi.org/10.1016/j.baae.2024.06.004
Parsons, S. E. y Frank, S. D. (2019). Urban tree pests and natural enemies respond to habitat at different spatial scales. Journal of Urban Ecology, 5, juz010. https://doi.org/10.1093/jue/juz010
Pascacio-Villafán, C. y Cohen, A. C. (2023). How rearing systems for various species of flies benefit humanity. Insects, 14, 553. https://doi.org/10.3390/insects14060553
Persson, A. S., Hederström, V., Ljungkvist, I., Nilsson, L. y Kendall, L. (2023). Citizen science initiatives increase pollinator activity in private gardens and green spaces. Frontiers in Sustainable Cities, 4, 1099100. https://doi.org/10.3389/frsc.2022.1099100
Ramírez-Restrepo, L. R. y Halffter, G. (2013). Butterfly diversity in a regional urbanization mosaic in two Mexican cities. Landscape and Urban Planning, 115, 39–48.
https://doi.org/10.1016/j.landurbplan.2013.03.005
Rocha, E. A. y Fellowes, M. D. E. (2018). Does urbanization explain differences in interactions between an insect herbivore and its natural enemies and mutualists? Urban Ecosystems, 21, 405–417. https://doi.org/10.1007/s11252-017-0727-5
Rocha-Ortega, M. y Castaño-Meneses, G. (2015). Effects of urbanization on the diversity of ant assemblages in tropical dry forests, Mexico. Urban ecosystems, 18, 1373–1388. https://doi.org/10.1007/s11252-015-0446-8
Roguz, K., Chiliński, M., Roguz, A. y Zych, M. (2023). Pollination of urban meadows. Plant reproductive success and urban-related factors influencing frequency of pollinators visits. Urban Forestry and Urban Greening, 84, 127944. https://doi.org/10.1016/j.ufug.2023.127944
Ruiz-Montoya, L., Alias, V. L. A., Alias, V. D. J., Guillén, D. T. A. y de la Mora, E. L. F. (2014). Diversidad y distribución de familias de insectos en el Cerrito de San Cristóbal. En L.Ruiz-Montoya (Coord.), Diversidad biológica y enriquecimiento florístico del Cerrito de San Cristóbal (pp. 39–56). El Colegio de la Frontera Sur.
Rzedowski, J. (1978). Vegetación de México. México D.F.: Limusa.
Schmidt, O., Schmidt S., Häuser, C., Hausmann, A. y Van Vu, L. (2019). Using Malaise traps for collecting Lepidoptera (Insecta), with notes on the preparation of Macrolepidoptera from ethanol. Biodiversity Data Journal, 7, e32192. https://doi.org/10.3897/BDJ.7.e32192
Seymoure, B. M. (2018). Enlightening butterfly conservation efforts: the importance of natural lighting for butterfly behavioral ecology and conservation. Insects, 9, 22. https://doi.org/10.3390/insects9010022
Sire, L., Yáñez, P. S., Wang, C., Bézier, A., Courtial, B., Cours, J. et al. (2022). Climate-induced forest dieback drives compositional changes in insect communities that are more pronounced for rare species. Communications Biology, 5, 57. https://doi.org/10.1038/s42003-021-02968-4
Start, D., Barbour, M. A. y Bonner, C. (2020). Urbanization reshapes a food web. Journal of Animal Ecology, 89, 808–816. https://doi.org/10.1111/1365-2656.13136
Tavares-Brancher, K. P., Graf, L. V., Ferreira-Júnior, W. G., Faria, L. D. B. y Zenni, R. D. (2024). Plant-pollinator interactions in the neotropics are affected by urbanization and the invasive bee Apis mellifera. Journal of Insect Conservation, 28, 251–261. https://doi.org/10.1007/s10841-024-00547-6
Triplehorn, C. A. y Johnson, N. F. (2005). Borror and Delong’s introduction to the study of insects. 7th Edition. Independence, KY: Cengage Learning.
Trivellone, V., Forte, V., Filippin, L. y Dietrich, C. H. (2021). First records of the North American leafhopper Gyponana mali (Hemiptera: Cicadellidae) invading urban gardens and agroecosystems in Europe. Acta Entomologica Musei Nationalis Pragae, 61, 213–219. https://doi.org/10.37520/aemnp.2021.011
Vergnes, A, Pellissier, V, Lemperiere, G, Rollard, C. y Clergeau, P. (2014). Urban densification causes the decline of ground-dwelling arthropods. Biodiversity and Conservation, 23, 1859–1877. https://doi.org/10. 1007/s10531-014-0689-3
Wagner, D. L., Grames, E. M., Forister, M. L., Berenbaum, M. R. y Stopak, D. (2021). Insect decline in the Anthropocene: Death by a thousand cuts. Proceedings of the National Academy of Sciences, 118, e2023989118. https://doi.org/10.1073/pnas.2023989118
Whittaker, R. H. (1972). Evolution and measurement of species diversity. Taxon, 21, 213–251. https://doi.org/10.2307/1218190
Wonderlin, N. E., Rumfelt, K. y White, P. J. (2019). Associations between nocturnal moths and flowers in urban gardens: evidence from pollen on moths. The Journal of the Lepidopterists’ Society, 73, 173–176. https://doi.org/10.18473/lepi.73i3.a6

Citizen science suggests decreased diversity of insects in Mexico, a megadiverse country
Datos de ciencia ciudadana sugieren un decremento en la diversidad de insectos de México, país megadiverso
Jorge Soberon *
University of Kansas, Biodiversity Institute, Dyche Hall, 1345 Jayhawk Blvd, Lawrence, KS 66045, USA
*Corresponding author: jsoberon@ku.edu (J. Soberon)
Received: 09 July 2025; accepted: 26 August 2025
Abstract
An analysis of iNaturalist data on several taxonomic groups of insects in Mexico is presented. A decreasing trend was observed in species diversity per year for 4 families of butterflies, bumblebees, and dragonflies and damselflies. Analyses were performed on several potential vegetation types (sensu Rzedowsky), and the roles of deforestation and pesticide use on the identified trends were explored. Challenges in using unsystematic data to estimate trends are discussed, and several hypotheses are provided to explain the results.
Keywords: Butterflies; Bumblebees; Damselflies; Dragonflies, iNaturalist
Resumen
Se presenta un análisis de datos de iNaturalist sobre varios grupos taxonómicos de insectos en México. Se observó una tendencia decreciente en la diversidad de especies por año para 4 familias de mariposas, y para abejorros y libélulas. Se realizaron análisis sobre varios tipos potenciales de vegetación (sensu Rzedowsky) y se exploró el papel de la deforestación y el uso de pesticidas en las tendencias identificadas. Se discuten los desafíos del uso de datos no sistemáticos para estimar tendencias y se presentan varias hipótesis para explicar los resultados.
Palabras clave: Mariposas, Abejorros; Caballitos del diablo; Libélulas; NaturaLista
Introduction
Evidence indicates a decrease in insect populations in many countries (Edwards et al., 2025; Hallmann et al., 2017). This finding is worrisome for many reasons, including that insects are key components of ecosystems and provide societies with important ecosystem services, such as pollination (Potts et al., 2010). Moreover, insects have substantial but largely unappreciated cultural importance (Duffus et al., 2021), not only worldwide but particularly in countries such as Mexico, where insects have culinary uses (Ramos-Elorduy & Viejo-Montesinos, 2007), have been important for ancestral cultures (Beutelspacher, 1989), and have economic and societal value (Ayala et al., 2012; Rogel-Fajardo et al., 2011).
Most detailed evidence of the decline in insects has come from countries in temperate zones that have developed formal monitoring schemes (Streitberger et al., 2024; Thomas, 2005). In contrast, tropical regions are less well studied (Sánchez-Herrera et al., 2024), and the existing evidence is contradictory (Bonadies et al., 2024; Boyle et al., 2025; Wagner et al., 2021). For instance, studies on Hemiptera (Lucas et al., 2016) and on saturniid moths (Basset et al., 2017) have indicated no trends in Barro Colorado Island, Panama. Similarly, in Veracruz, Mexico, well monitored fruit flies have shown no trends (Aluja et al., 2012; Ordano et al., 2013). In contrast, decreases in saturniid larvae have been reported in Costa Rica (Salcido et al., 2020), and declines in arthropod biomass have been reported in Puerto Rico and, on the basis of a few data points, in Chamela, Mexico (Lister & García, 2018). The monarch butterfly, perhaps the best monitored insect species in Mexico, has shown consistent decreases in its wintering aggregations (Thogmartin et al., 2017; Vidal & Rendón-Salinas, 2014; Zylstra et al., 2021).
Because of its history, climate, topography, and cultural milieu (Ramamoorthy et al., 1993), Mexico is among the world’s megadiverse countries (Mittermeier et al., 1997). Therefore, assessing the trends in insect populations in Mexico should be prioritized. Unfortunately, long-term insect monitoring in Mexico is rare. Although Mexico has a long history of entomological research, including many collections and hundreds of publications (Michán & Llorente, 2002), monitoring has been limited to only a few species. Although the reasons for the lack of national monitoring schemes like those existing in other countries should be determined, this study does not attempt to do so. It takes as a premise that, in Mexico, just a few systematically obtained insect time series of more than 2-3 years long are available. This study is aimed at estimating insect biodiversity trends in Mexico, despite the absence of systematic monitoring efforts.
Systematic monitoring results are compiled in several worldwide time series databases, such as the Living Planet Index (Almond et al., 2020) and the Global Population Dynamics database (NERC Centre for Population Biology, 1999). Unfortunately, these databases have sparse insect information and contain no data for Mexico. Another possibility is using so-called citizen science (CS) data (Cohn, 2008), which, although opportunistic and unsystematic, is often abundant. Data on insects collected by non-professionals have been used to estimate phenology and distributions (Soroye et al., 2018). However, using such data to estimate population trends is challenging, as discussed below.
In Mexico, perhaps the most comprehensive CS initiative is iNaturalist (known as Naturalista in Mexico). iNaturalist began its operations in Mexico in 2008, although in 2013 the initiative came under the leadership of the national biodiversity agency, Conabio, under the name of Naturalista (Macías & Freire, 2017) and obtained funding from the Slim Foundation. Therefore, in Mexico, iNaturalist began in earnest in 2013. Despite this relatively late start, Mexico is the third country in amount of data (Mason et al., 2025) and it contains more than 80,000 records tagged as “research level” for 3 families of butterflies, and the damselflies, dragonflies, and bumblebees. This substantial information may be used to assess trends. However, CS data must be corrected for biases, of which are many (Crall et al., 2011). Specifically, in Mexico, the number of observers (and thus of observations) in iNaturalist increases each year (Table S1), and this bias should be considered when using such data.
Indeed, a major problem in using opportunistic CS data to estimate trends is correcting for biases in recording efforts (Di Cecco et al., 2021). Several methods can be used to address this problem (Isaac et al., 2014; Outhwaite, 2019; Tang et al., 2021). One of the simplest methods is correcting bias by obtaining the quotient of the metric used to report biodiversity to some measure of the effort invested in a locality, for a given period. What is “effort,” and how can it be measured in iNaturalist data? Collection effort is difficult to define but can be described in terms of: 1) the time spent collecting, 2) the method of collection and number of collectors, or 3) the number of specimens or species observed (Gulland, 1969; Willott, 2001). iNaturalist data allows for extraction of a measure of time (number of monthly observations in a year), but data quality (beyond the “research” tag, which refers to the reliability of the name assigned to the species), remains unreported, and worse, in the case of iNaturalist, this quality is known to change (Di Cecco et al., 2021). Di Cecco (2021) has suggested that, in iNaturalist, observers with at least 2 observations are more reliable than those with just 1 observation. Therefore, as a measure of effort, this study used the number of observers with 2 or more observations. As biodiversity measures it is used the number of species, and the number of observations, pooled for spatial units and year. Two indices are then calculated: number of species/effort, and number of records/effort.
Ordinary regressions of metrics against time often experience problems of autocorrelated errors and non-equal variances (heteroskedasticity). These are characteristic of time series (Shumway & Stoffer, 2005) and must be accounted for. One method of addressing the complexities of analysis of count time series data is using a package such as “trim” (in the R platform), which assumes a Poisson model for the underlying data (Pannekoek, 1998). This approach corrects for the autocorrelation of errors and for heteroskedasticity. Trim has frequently been used for European (van Strien et al., 2019) and tropical American (Novoyny & Basset, 2000) data, but the key assumption of count data (a discrete scale) complicates analysis of continuous-scale indices, or data including many non-occurrences, because the software is sensitive to the presence of zeroes, or NAs, in the data.
Another possibility is estimating whether a significant trend exists in the data, by using a non-parametric Mann-Kendall test (Lyubchich et al., 2013). An ordinary least squares linear regression (OLS) of metric against time is first performed, and the existence of trends (linear or monotonic) is subsequently determined. This method uses the sign of the slope in the OLS, and the significance is tested with the Mann-Kendall test.
Additional methods can be used, such as, for a single species, the logit of the probability of occupancy of a cell, on a time unit (van Strien et al., 2019) and fit a generalized linear model of predictors, by using the length of the list of species as a measure of effort (Szabo et al., 2010). Then several single species regressions can be combined in an index (van Strien et al., 2019). One problem with this approach is that generalized linear modeling is based on an assumption of independence of errors, which might be violated in a time series.
A statistically more sophisticated modification of the above idea is reporting the proportion of occupied sites under a hierarchical model that separates the actual presence from the act of observation (Outhwaite, 2019). Although apparently very rigorous, this approach has its own problems, including the need to define an appropriate model for the “present” and “observer” components, and the need to have replicated visits to the same site within the same season (van Strien et al., 2019).
Another possibility is using generalized least squares (GLS) regressions, which allow for autocorrelated errors and heteroskedasticity. The R package “nlme” implements this technique. This method can fit an ordinary regression of the index against covariates such as time, and another regression including autocorrelation with power variance decay in its model. Subsequently, the 2 models can be compared with the Akaike criterion, and the best model can be retained. This option was used here, with the simplest ARIMA model with lag = 1 as a model of correlated errors.
This study further assessed whether any existing trends might have differed for different ecological regions of Mexico. A variety of subdivisions of Mexico have been suggested, according to different ecological perspectives, at different spatial resolutions (Anonymous, 1997; Challenger & Soberón, 2012; Miranda & Hernández, 1963; Olson et al., 2001). Here, Rzedowsky’s potential vegetation types were used (Rzedowsky, 1986). Although coarse-grained, these types are based primarily on straightforward floristic criteria, are well known in Mexico, and have a small number of categories.
An important caveat in using CS data is that species that are difficult to identify by sight should be avoided. This work focused on 3 families of butterflies (Papilionidae, Pieridae, and Nymphalidae), with 316 names (skippers and the smallest families in the Papilionoidea were excluded); 23 names for bumblebees; and 293 names for the Odonata (both Zygoptera and Anysoptera). In addition, as a comparison, data on 307 names for Solanaceae were included. The numbers of names (without proper taxonomic validation by experts), as reported by iNaturalist, are listed in Table 1.
For the butterflies, although using species identified as indicators of “conservation” status (Orta et al., 2022) would have been interesting, most species identified by these authors as indicators had only a few records in the iNaturalist database. Therefore, the analysis was performed not by individual species, but by pooling all the data in the 3 families of butterflies, all the dragon and damselflies, and all the bumblebees.
For obtaining uncertainty bands, grids of hexagons covering the territory of Mexico were defined at several resolutions (Fig. 1). For a given year and taxonomic group, the means and variances over hexagons were determined. Each unique combination of year and hexagon defined an “event,” and thus the abundance metrics were: 1) the number of observations per event (cumulative monthly observations); and 2) the number of different species per event. As a measure of effort, the total number of different observers with at least 2 observations in each “event” was used. The final index was the average over all hexagons with at least 1 record, for a given year, of the number of observations or the number of species, per observer.
Table 1
Numbers of scientific names for the different taxonomic groups in the 4 most visited potential vegetation classes. The butterflies are the Papilionidae (swallowtails), Pieridae (sulfurs), and Nymphalidae (brushfoots).
| All Mexico | Xerophytic Shrub | Pine Oak Forest | Grasslands | Tropical Deciduous Forest | |
| Butterflies | 413 | 153 | 220 | 220 | 198 |
| Odonata | 292 | 185 | 187 | 105 | 184 |
| Bombus | 23 | 16 | 20 | 9 | 14 |
| Solanaceae | 307 | 176 | 210 | 111 | 153 |
Changing the hexagon area might potentially change the results. This problem, described as the “modifiable areal unit problem,” has been long known to geographers (Openshaw, 1984). Fortunately, in this case, the qualitative results were not affected by the resolution of the hexagons (data correlations among resolutions always exceeded 0.7). Consequently, only the analysis using the largest (2 degrees) hexagons (n = 81) is reported.
The literature has suggested that the decrease in insect abundance has been due to: 1) increased use of pesticides, 2) decreased habitat area (or increased transformed land area), and 3) climate change. At the scale of the whole country, regressions of data versus time series of pesticide use and deforestation rates are reported.

Materials and methods
CS data are not ideally suited to the estimation of trends, primarily because of the biased and uneven methods of sampling sites, times, and species. This work used 1 of the 3 methods proposed by Isaac et al. (2014): correcting the reported number of sightings according to a measure of effort. iNaturalist data were downloaded from the Global Biodiversity Information Network (GBIF), as detailed in Table 2.
Data were divided into subsets (keeping records with coordinates) for the 4 largest families in the Papilionoidea: Papilionidae (5,583 records), Pieridae (29,994 records), Nymphalidae (20,145 records), and Lycaenidae (2,326 records). The Lycaenidae was removed from the analysis because many species are relatively difficult to determine visually. Data for the genus Bombus (bumblebees, 6,543 records) and the 2 suborders of the Odonata (the Zygoptera, 12,772 records, and the Anisoptera, 24,003 records) were also downloaded. For comparison purposes, observations of the nightshade family, the Solanaceae (39,014 records), were downloaded. The number and positions of every observation in Mexico are presented in figure 2.
Table 2
Digital Object Identifiers (DOIs) from GBIF for the datasets used in the work.
| Taxon | GBIF DOI | iNaturalist Records | Unique names |
| Nymphalidae | doi.org/10.15468/dl.qta4zp | 20,145 | 223 |
| Papilionidae | doi.org/10.15468/dl.uu4unc | 5,583 | 13 |
| Pieridae | doi.org/10.15468/dl.zug4ee | 29,994 | 80 |
| Lycaenidae | doi.org/10.15468/dl.xgad6g | 2,326 | 97 |
| Odonata | doi.org/10.15468/dl.atdkfz | 36,775 | 292 |
| Anisoptera | 24,003 | 163 | |
| Zygoptera | 12,772 | 129 | |
| Bombus | doi.org/10.15468/dl.c6h4jz | 6,543 | 23 |
| Solanaceae | doi.org/10.15468/dl.597nj5 | 39,014 | 307 |
Data tagged as “research quality” in the downloaded GBIF data were retained, and basic data cleaning was performed to keep the coordinates inside Mexico. No attempt was made to correct for outdated taxonomy or other known issues present in aggregator data (Chapman, 2005).
Data can be organized as time series, by pooling the observations in a year. This method has a drawback of potentially missing seasonality; however, pooling by month produces tables that are too sparse and therefore are difficult to analyze. To include some measure of uncertainty in the trends, the averages of the calculated indices over all non-empty (i.e., with at least 1 observation) hexagons of 2 degrees of surface were determined, and its standard error calculated.
Two indices were used: different_species/observer and records/observer. The first is a measure of diversity, whereas the second is a measure of abundance. Findings for both are reported. “Observers” refers to the number of observers with at least 2 registered observations.
To summarize trends, a useful statistic may be the slope of a linear model of index as a function of time, which requires regressions of index vs. year. However, as previously discussed, the errors in many time series are not independent, and the equal variance assumption of ordinary least squares is also often violated. If uncorrec-
ted, these problems interfere with rigorous calculations of probability under a null hypothesis (McShane et al., 2019). Among the many methods for addressing this problem, generalized least squares regressions (Baillie & Kim, 2018), which enable inclusion of an autoregressive structure of correlations and violations of homoscedasticity, were chosen herein. Two models were fitted to the data: an ordinary linear least squares, and a first order auto-regressive, moving average model (ARIMA) (Shumway & Stoffer, 2005) allowing for heteroskedasticity. The 2 models were compared with an ANOVA (Fox & Weisberg, 2019), and the most likely model (based on the Akaike criterion; see Fox & Weisberg, 2019) was used. This process permitted to obtain, in a rigorous way, the probability for the observed slope values, under a null hypothesis of a slope equal to zero. Reporting the “significance” of slopes has been substantially criticized (McShane et al., 2019). Therefore, the probability (rather than the “significance”) of the slope, based on the assumption of a null model of no trend, is reported. Very small probabilities are highlighted.
The regressions included 2 possible causal factors: forest loss and use of pesticides. The deforestation rate was obtained from the Global Forest Watch website (Sims et al., 2024) with a threshold of 30% of forest cover, as recommended by Sims et al. (2024). This dataset has maintained methodological consistency (Hansen et al., 2013) and therefore is preferable to the INEGI Series (Gebhardt et al., 2015). Agrochemical use was determined as the amount of pesticides used per hectare of cropland, as reported on the FAO Web site. The data came from government reports https://www.fao.org/faostat/en/#data/RP. A discussion of the FAO dataset’s strengths and problems has been provided by Shattuck et al. (2023).
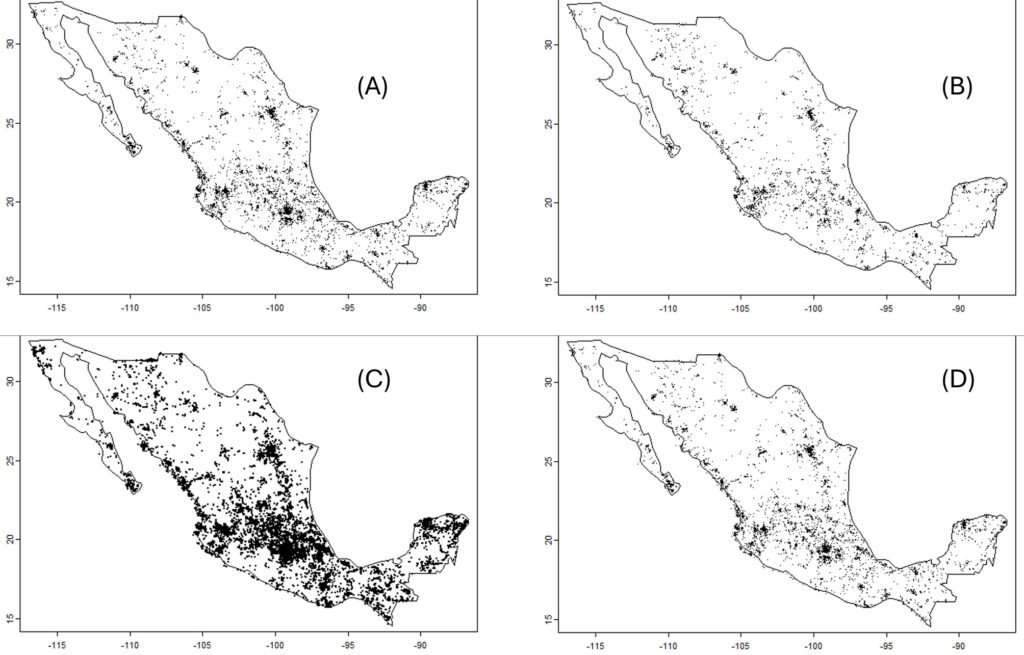
Because the probability of the observed values of the slope of the index of diversity per unit of effort vs. time, under a null hypothesis of 0 slope, was small in most cases, the regression was assumed to remove the time trend, and factors affecting just the residuals were searched for. That is, the residual of the index vs. time regressions was regressed against 2 predictors: deforestation rate and use of pesticides. The results are shown in the Supplementary materials.
To aggregate by “biome,” the subdivision of the Potential Vegetation of Mexico (Rzedowsky, 1986) was selected. A shapefile of Rzedowsky’s map at 1:4,000,000 scale, available at Conabio Geoportal, is produced by Instituto de Geografía, UNAM México. This map was used to pool the iNaturalist records according to potential vegetation, by using the 4 categories with the highest number of iNaturalist reports.
An informal survey was circulated among scientists working in 3 major ecology research centers in Mexico (INECOL, Veracruz, Instituto de Ecología, UNAM, and Ecosur, Chiapas). A total of 37 questionnaires were sent with Qualtrics. The questions are provided in the Supplementary materials. The main data tables and R code are openly available (Creative Commons CC0: 1) at https://github.com/jsoberon/iNaturalistInsectsMexico
Results
The informal questionnaire received 27 responses out of 37 requests. Among the respondents, 84% stated that they have observed a decrease in the number of insects either in streetlights in villages, or in the windshields or radiators of field vehicles. Although these answers lacked statistical rigor, they suggested a widespread perception among field biologists in Mexico that insect populations are becoming smaller.
The iNaturalist data provided a more nuanced picture. Before examining the trends in biodiversity indices, basic data were analyzed. Indeed, both the number of species and the number of observers (with more than 2 observations) increased (Fig. 3).
The numbers of observed species and observers both increase over time. The increased number of observers introduced an important bias in the data, given that more species (or more individuals) would reasonably be expected to be reported if more observers were present. However, although the diversity of insects appeared to be decreasing, the evidence of a decrease in abundance was unclear (Fig. 4; Tables 3, 4).
Diversity per unit effort appeared to decrease (Table 3). However, the trends in the abundance (observations/number observers) were either positive or indistinguishable from 0 (Table 4). Box plots of the slopes of the regressions for the 2 indices (species, and observations) are shown in figure 5.
The above results suggest that diversity is decreasing, but abundance is stable. This finding is inconsistent with the informal perceptions of field biologists (as indicated by the questionnaire), most of whom perceived diminished insect abundance. Among the few insect species whose abundance in Mexico has been monitored systematically, Danaus plexippus (monarch butterfly) populations are decreasing (Vidal & Rendón-Salinas, 2014; Zylstra et al., 2021), whereas Anastrepha fruit fly populations appear to be stable (Aluja et al., 2012; Ordano et al., 2013). Comparing these 2 cases is challenging, because monarch butterflies are affected by a variety of factors occurring on a continental scale, whereas fruit flies might be affected primarily by local factors.
Might the negative trend in diversity correlate with predictors often associated with insect loss? Forest cover, as measured via remote sensing over 15 years (Hansen et al., 2013), is decreasing in Mexico (Supplementary materials). Pesticide use per hectare of crop, as reported by the FAO, increased until 2018, when the FAO database indicated an abrupt decrease (Supplementary materials). The causes of this decrease, if real, are unknown; however, after the COVID-19 pandemic, Mexico’s primary sector experienced a marked decrease in activity (Sánchez et al., 2022), which may explain a drop in the use of agrochemicals. Regressions of the residuals of the diversity/effort vs. time models against 2 predictors, deforestation rate and pesticide use per hectare, were not associated with small probabilities of an H0 of 0 slope (Supplementary materials). Consequently, the data did not provide evidence that negative slopes in insect diversity were due to pesticide use or deforestation.
Finally, for the major taxonomic groups, the slopes of the generalized least squares, in the first 4 potential vegetation types according to Rzedowsky (1986) were most negative for bumblebees in tropical deciduous forest, followed by pine-oak forest and xerophytic shrub. For the butterflies, the most negative slope was in pine-oak forest, followed by tropical deciduous forest and xerophytic shrub (Supplementary materials: Table S3). In the case of the Solanaceae, a group included for comparison purposes, the slope is only negative in the grasslands vegetation type.
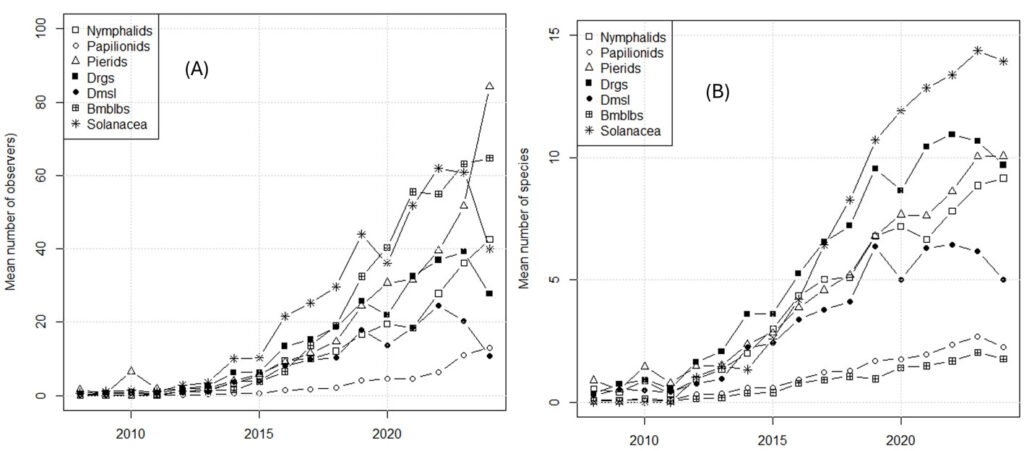
Table 3
Regression analysis (generalized least squares) of diversity/observer vs. time in the iNaturalist data, for the main taxonomic groups. The analysis was performed over the mean values in hexagons of 2 degrees of resolution. With the exception of the Zygoptera, for which the first order autoregressive model did not converge, the ordinary least squares regression did not significantly differ with respect to models with autocovariance and heteroskedasticity. Consequently, the table shows the slope of ordinary linear models of different_species/effort with respect to time. The probabilities of the obtained values under a null hypothesis of slope of zero were very small, with the exception of the dragonflies and swallowtails (Fig. 2).
| Taxon | Species, 2 degrees | |||
| Slope | p | Model | n | |
| Bombus | -0.0428 | 0.0000237 | OLS | 6,543 |
| Anisoptera | -0.0206 | 0.00155 | OLS | 24,003 |
| Zygoptera | 0.0003 | 0.942 | OLS_NO_CNV | 12,772 |
| Nymphalidae | -0.0372 | 0.000000242 | OLS | 20,145 |
| Papilionidae | -0.0108 | 0.104 | OLS | 5,583 |
| Pieridae | -0.0247 | 0.000202 | OLS | 29,994 |
Table 4
Regression analysis (generalized least squares) of records/observer vs. time in the iNaturalist data, for the main taxonomic groups. The data were averaged over hexagons of 2 degrees of resolution. Except for the Zygoptera, for which a first order autoregressive model was used, the ordinary least squares regression did not significantly differ with respect to models with autocovariance and heteroskedasticity. Consequently, the table shows the slope of ordinary linear regressions of number_of_records/effort with respect to time. Notably, every regression had a positive, low probability slope.
| Taxon | Slope | p | Model | n |
| Bombus | 0.0446 | 0.00000766 | OLS | 6,543 |
| Anisoptera | 0.0201 | 0.00649 | OLS | 24,003 |
| Zygoptera | 0.0366 | 6.86E-08 | ARIMA | 12,772 |
| Nymphalidae | 0.0089 | 0.034 | OLS | 20,145 |
| Papilionidae | 0.0669 | 0.00000176 | OLS | 5,583 |
| Pieridae | 0.0154 | 0.0000984 | OLS | 29,994 |

Discussion
The results show a tendency to decrease the number of species with time, for the insects, and a much less marked negative trend for the Solanaceae. This suggests that CS data does capture some sort of biological signal in the data. However, a diminishing trend of diversity, together with a stable pattern of abundance, are compatible with several hypotheses. One entirely biological hypothesis is that insect diversity, but not abundance, is decreasing. If the rarest species are disappearing, then the country is homogenizing (McKinney & Lockwood, 1999). Thus, Mexico’s highly diverse and unique insect biodiversity is slowly being replaced by a more homogeneous, more cosmopolitan set of species. This rather alarming possibility, supported by the CS data, must be more directly assessed in the field.
Another explanation for the observed negative trend in insect species numbers might be that, over time, observers have reached the asymptote of the total number of species available to be observed. Since the total number of species in any given area is probably roughly constant, with sufficient effort, no more than that constant number can be reported; however, if the number of observers is increasing, a negative trend in the index of species/observers would result. The total number of species in the database, for each taxonomic group, is shown in Table 1. The average number of species reported per hexagon was well below that total (Supplementary materials: Table S4), thus suggesting that a saturation effect was not present, and the results presented here indeed indicate a decreasing trend in insect diversity. This complex point is discussed at more length in the Supplementary materials.
Finally, the negative trend is also compatible with a hypothesis regarding the quality of iNaturalist observers in which the number of observers has increased, as indicated by the data, whereas the observers’ discrimination ability or interests might have changed over time, perhaps because they focused on common species. Unfortunately, the very nature of the information in CS data makes assessing this effect very difficult. This aspect essentially describes the main problem with using unstructured CS data: because the methods are not standardized, any trend in the data might be explained by a trend in the behavior of the observers.
What explanations can be deduced for the absence of trends in the number of observations/effort? One possibility is that the presence of more observers simply resulted in more observations, and the number of observations and observers with more than 2 observations are roughly proportional. This means that the lack of trend could be an artifact of the data.
These results should be considered as hypotheses to be examined through more direct methods. Nonetheless, the results strongly suggest decreasing numbers of species in butterflies (important from a cultural perspective and perhaps a pollination perspective), bumblebees (important as pollinators), and Odonata (important as insect predators and as indicators). Therefore, the biodiversity of some of the most important and underappreciated groups of species in Mexico appears to be decreasing. If confirmed, this result would be highly alarming. Indeed, insects are key components of ecosystems (Noriega et al., 2018). Although for most insect species in Mexico we do not have direct documentation of their role, or of the economic and cultural value of their services, we have substantial indirect evidence of their importance as pollinators (Ashworth et al., 2009), as natural enemies of agricultural pests (López et al., 1999; Aluja et al., 2014), and as potential for non-conventional agri-business (López-Gutierrez et al., 2023). If substantiated, the decrease we report should be a major cause of alarm for Mexicans.
What might be causing a decrease? In a study in Europe (Schuch et al., 2012), in which a similar decrease in diversity was reported in a family of bugs of agricultural importance, a concomitant loss of non-agricultural habitat for the insects was reported. The study was conducted at the species level, and the authors argue that the more specialized, less tolerant species are those disappearing because of agricultural expansion. Again, monitoring using standardized procedures is required to test this idea.
Climate change is often cited as a cause of population decline in insects. However, climate change is a long-term phenomenon that occurs at the scale of many decades. To demonstrate climate change as a factor affecting population size, modeling or documentation of the effect of mean and variance in climatic variables on long-term population time series is necessary (Batalden et al., 2014; Boggs, 2016). The data used in this work is not appropriate for this purpose.
The results suggested that negative trends might not be identical among ecological regions. However, interestingly, the iNaturalist data indicated that pine-oak forest, xeric shrub, and tropical deciduous forest might be hotspots of diversity loss. This finding is somewhat surprising, given the widespread concern regarding tropical rainforests. Of course, the results may be due to the scarcity of data for tropical wet vegetation types.
CS data exists in substantial and growing amounts. It is a very valuable source of data. However, as with any data, it contains biases that are sometimes difficult to remove. The unavoidable conclusion is that Mexico must crucially invest in countrywide insect monitoring schemes based on systematic methods. Several approaches could be used. The first is improving CS schemes, providing training, and applying standard protocols, as performed in Canada, the USA, and many European countries (Streiter et al., 2024). This approach might be useful only for conspicuous, easily identifiable species, yet it markedly influences public environmental awareness and therefore should be maintained (Dickinson et al., 2012). Several methods using advanced technologies include computer vision, bioacoustics, and metagenomics can also be used (Van Klink et al., 2022). For bats, monitoring is already underway in Mexico (Zamora-Gutiérrez et al., 2020). Adoption of high technology methods would require funding, training, and substantial analytical capacity.
Regardless of the method chosen, in Mexico, the fourth most biologically diverse country on the planet, monitoring as many biodiversity components as possible is critical, and insects, “the little things that run the world” in the words of E. O. Wilson, appear to be disappearing very rapidly. Societies need to pay attention.
Acknowledgements
I am very grateful to Luis Eguiarte and Rodrigo Medellín, of the Mexican Instituto de Ecología, for their detailed and positive criticism of the methods I used. Their feedback led to several changes in the data analysis methods. Exequiel Ezcurra, of the University of California at Irvine, and Carlos Martinez, formerly of the University of Wyoming, also made very helpful statistical suggestions. My students Jennifer Ramos and Anahí Quezada helped me download and organize the data and patiently discussed the project with me. I gratefully thank the field ecologists in Mexico who took the time to respond to the questionnaire regarding their experiences with insects in the field.
References
Almond, R. E., Grooten, M., & Peterson, T. (2020). Living planet report 2020-Bending the curve of biodiversity loss. Gland, Switzerland: World Wildlife Fund.
Aluja, M., Ordano, M., Guillén, L., & Rull, J. (2012). Understanding long-term fruit fly (Diptera: Tephritidae) population dynamics: implications for areawide manage-
ment. Journal of Economic Entomology, 105, 823–836. https://doi.org/dx.doi.org/10.1603/EC11353
Aluja, M., Sivinski, J., Van Driesche, R., Anzures-Dadda, A., & Guillén, L. (2014). Pest management through tropical tree conservation. Biodiversity and Conservation, 23, 831–853. https://doi.org/10.1007/s10531-014-0636-3
Anonymous. (1997). Ecological regions of North America. Towards a common perspective. Montreal: Comisión de Cooperación Ambiental de América del Norte.
Ashworth, L., Quesada, M., Casas, A., Aguilar, R., & Oyama, K. (2009). Pollinator-dependent food production in Mexico. Biological Conservation, 142, 1050–1057. https://doi.org/10.1016/j.biocon.2009.01.016
Ayala, R., González, V. H., & Engel, M. S. (2012). Mexican stingless bees (Hymenoptera: Apidae): diversity, distribution, and indigenous knowledge. Pot-honey: a legacy of stingless bees. New York: Springer. https://doi.org/10.1007/978-1-4614-4960-7_9
Baillie, R. T., & Kim, K. H. (2018). Choices between OLS with robust inference and feasible GLS in time series regressions. Economics Letters, 171, 218–221. https://doi.org/10.1016/j.econlet.2018.07.036
Basset, Y., Lamarre, G. P., Ratz, T., Segar, S. T., Decaëns, T., Rougerie, R. et al. (2017). The Saturniidae of Barro Colorado Island, Panama: a model taxon for studying the long-term effects of climate change? Ecology and Evolution, 7, 9991–10004. https:// doi.org/10.1002/ece3.3515
Batalden, R. V., Oberhauser, K., & Peterson, A. T. (2014). Ecological niches in sequential generations of eastern North American Monarch butterflies (Lepidoptera: Danaidae): the ecology of migration and likely climate change implications. Environmental Entomology, 36, 1365–1373. https://doi.org/10.1603/0046-225x(2007)36[1365:enisgo]2.0.co;2
Beutelspacher, C. R. (1989). Las mariposas entre los antiguos mexicanos. Mexico D.F.: Fondo de Cultura Económica.
Boggs, C. L. (2016). The fingerprints of global climate change on insect populations. Current Opinion in Insect Science, 17, 69–73. https://doi.org/10.1016/j.cois.2016.07.004
Bonadies, E., Lamarre, G. P., Souto-Vilarós, D., Pardikes, N. A., Silva, J. A. R., Perez, F. et al. (2024). Population trends of insect pollinators in a species-rich tropical rainforest: stable trends but contrasting patterns across taxa. Biology Letters, 20, 20240170. https://doi.org/10.1098/rsbl.2024.0170
Boyle, M. J., Bonebrake, T. C., Dias da Silva, K., Dongmo, M. A., Machado-França, F., Gregory, N. et al. (2025). Causes and consequences of insect decline in tropical forests. Nature Reviews Biodiversity, 1, 315–331. https://doi.org/10.1038/s44358-025-00038-9
Challenger, A., & Soberón, J. (2012). Los ecosistemas terrestres, en Capital natural de México. Conocimiento Actual de la Biodiversidad. México D.F.: Conabio.
Chapman, A. (2005). Principles and methods of data cleaning-primary species and species-occurrence data. Copenhagen: Global Biodiversity Information Facility.
Cohn, J. P. (2008). Citizen science: Can volunteers do real research? Bioscience, 58, 192–197. https://doi.org/10.1641/B580303
Crall, A. W., Newman, G. J., Stohlgren, T. J., Holfelder, K. A., Graham, J., & Waller, D. M. (2011). Assessing citizen science data quality: an invasive species case study. Conservation Letters, 4, 433–442. https:// doi.org/10.1111/j.1755-263X.2011.00196.x
Di Cecco, G. J., Barve, V., Belitz, M. W., Stucky, B. J., Guralnick, R. P., & Hurlbert, A. H. (2021). Observing the observers: How participants contribute data to iNaturalist and implications for biodiversity science. Bioscience, 71, 1179–1188. https://doi.org/10.1093/biosci/biab093
Dickinson, J. L., Shirk, J., Bonter, D., Bonney, R., Crain, R. L., Martin, J. et al. (2012). The current state of citizen science as a tool for ecological research and public engagement. Frontiers in Ecology and the Environment, 10, 291–297. https://doi.org/10.1093/biosci/biab093
Duffus, N. E., Christie, C. R., & Morimoto, J. (2021). Insect cultural services: how insects have changed our lives and how can we do better for them. Insects, 12, 377. https://doi.org/10.3390/insects12050377
Edwards, C. B., Zipkin, E. F., Henry, E. H., Haddad, N. M., Forister, M. L., Burls, K. J. et al. (2025). Rapid butterfly declines across the United States during the 21st century. Science, 387, 1090–1094. https://doi.org/10.1126/science.adp4671
Fox, J., & Weisberg, S. (2019). Nonlinear regression, nonlinear least squares, and nonlinear mixed models in R. In J. Fox, & Weisberg, S. (Eds.), An R companion to applied regression, 3rd Ed. (pp. 1–31). Los Angeles: Sage Publications.
Gebhardt, S., Maeda, P., Wehrmann, T., Argumedo-Espinoza, J., & Schmidt, M. (2015). A proper land cover and forest type classification scheme for Mexico. The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, 40, 383–390. https://doi.org/10.5194/isprsarchives-XL-7-W3-383-2015
Gulland, J. A. (1969). Manual of methods for fish stock assessment. Part 1. Fish Population Analysis. Rome: Fishery Resources and Exploitation Division, FAO.
Hallmann, C. A., Sorg, M., Jongejans, E., Siepel, H., Hofland, N., Schwan, H. et al. (2017). More than 75 percent decline over 27 years in total flying insect biomass in protected areas. Plos One, 12, e0185809. https://doi.org/10.1371/journal.pone.0185809
Hansen, M. C., Potapov, P. V., Moore, R., Hancher, M., Turubanova, S. A., Tyukavina, A., D. et al. (2013). High-resolution global maps of 21st-century forest cover change. Science, 342, 850–853. https://doi.org/ 10.1126/science.1244693
Isaac, N. J., van Strien, A. J., August, T. A., de Zeeuw, M. P., & Roy, D. B. (2014). Statistics for citizen science: extracting signals of change from noisy ecological data. Methods in Ecology and Evolution, 5, 1052–1060. https://doi.org/10.1111/2041-210X.12254
Lister, B. C., & García, A. (2018). Climate-driven declines in arthropod abundance restructure a rainforest food web. Proceedings of the National Academy of Sciences, 115, E10397–E10406. https://doi.org/10.1073/pnas.1722477115
López-Gutiérrez, S., Pozo, C., Muñoz, D. E. R., & Baltazar, E. B. (2023). The live butterfly trade as bio-business in southern Mexico. Journal of Rural and Community Development, 18, 131–152.
López, M., Aluja, M., & Sivinski, J. (1999). Hymenopterous larval–pupal and pupal parasitoids of Anastrepha flies (Diptera: Tephritidae) in Mexico. Biological Control, 15, 119–129. https://doi.org/10.1006/bcon.1999.0711
Lucas, M., Forero, D., & Basset, Y. (2016). Diversity and recent population trends of assassin bugs (Hemiptera: Reduviidae) on Barro Colorado Island, Panama. Insect Conservation and Diversity, 9, 546–558. https://doi.org/10.1111/icad.12191
Lyubchich, V., Gel, Y. R., & El-Shaarawi, A. (2013). On detecting non-monotonic trends in environmental time series: A fusion of local regression and bootstrap. Environmetrics, 24, 209–226. https://doi.org/ 10.1002/env.2212
Macías, C. G. V., & Freire, L. R. (2017). Plataforma Naturalista en México: herramienta de ciencia ciudadana. Índice, 12, 762.
Mason, B. M., Mesaglio, T., Barratt-Heitmann, J., Chandler, M., Chowdhury, S., Gorta, S. B. Z. et al. (2025). iNaturalist accelerates biodiversity research. Bioscience, 2025, biaf104. https://doi.org/10.1093/biosci/biaf104
McKinney, M. L., & Lockwood, J. L. (1999). Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends in Ecology & Evolution, 14, 450–453. https://doi.org/10.1016/S0169-5347(99)01679-1
McShane, B. B., Gal, D., Gelman, A., Robert, C., & Tackett, J. L. (2019). Abandon statistical significance. The American Statistician, 73, 235–245. https://doi.org/10.1080/00031305.2018.1527253
Michán, L., & Llorente, J. (2002). Hacia una historia de la Entomología en México. Publicaciones Especiales del Museo de Zoologia, UNAM, Mexico, 3, 3–52.
Miranda, F., & Hernández, E. (1963). Los tipos de vegetación de México y su clasificación. Botanical Sciences, 215, 29–179. https://doi.org/10.17129/botsci.1084
Mittermeier, R., Robles-Gil, P., & Mittermeier, C. (1997). Megadiversidad: los países biológicamente mas ricos del mundo. México D.F.: CEMEX.
NERC Centre for Population Biology, I. C. (1999). The global population dynamics database. https://knb.ecoinformatics.org/view/doi%3A10.5063%2FAA%2Fnceas.167.1
Noriega, J. A., Hortal, J., Azcárate, F. M., Berg, M. P., Bonada, N., Briones, M. J. et al. (2018). Research trends in ecosystem services provided by insects. Basic and Applied Ecology, 26, 8–23. https://doi.org/10.1016/j.baae.2017.09.006
Novoyny, V., & Basset, Y. (2000). Rare species in communities of tropical insect herbivores: pondering the mystery of singletons. Oikos, 89, 564. https://doi.org/10.1034/j.1600-0706.2000.890316.x
Olson, D. M., Wikramanayake, E. D., Burges, N. D., Powell, G. V. N., Underwood, E. C., D’Amico, J. A. et al. (2001). Terrestrial ecorregions of the world: a new map of life on earth. Bioscience, 51, 933. https://doi.org/10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2
Openshaw, S. (1984). The modifiable areal unit problem. Norwich: Geo Books.
Ordano, M., Guillén, L., Rull, J., Lasa, R., & Aluja, M. (2013). Temporal dynamics of diversity in a tropical fruit fly (Tephritidae) ensemble and their implications on pest management and biodiversity conservation. Biodiversity and Conservation, 22, 1557–1575. https://doi.org/10.1007/s10531-013-0468-6
Orta, C., Reyes-Agüero, J. A., Luis-Martínez, M. A., Muñoz-Robles, C. A., & Méndez, H. (2022). Mariposas bioindicadoras ecológicas en México. Acta Zoológica Mexicana, 38, 1–33. https://doi.org/10.21829/azm.2022.3812488
Outhwaite, C. L. (2019). Modelling biodiversity trends from occurrence records. London: UCL (University College London).
Pannekoek, J., & van Strien, A. (1998). TRIM 2.0 for Windows (TRends and Indices for Monitoring Data).
Potts, S. G., Biesmeijer, J. C., Kremen, C., Neumann, P., Schweiger, O., & Kunin, W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends in Ecology & Evolution, 25, 345–353. https://doi.org/10.1016/j.tree.2010.01.007
Ramamoorthy, T. R., Bye, R., Lot, A., & Fa, J. (1993). Biological diversity of Mexico: origins and distribution. Oxford: Oxford University Press. https://doi.org/ 10.1007/bf02908211
Ramos-Elorduy, J., & Viejo-Montesinos, J. (2007). Los insectos como alimento humano: breve ensayo sobre la entomofagia, con especial referencia a México. Boletín de la Real Sociedad Española de Historia Natural Sección Biológica, 10, 61–84.
Rogel-Fajardo, I., Rojas-Lopez, A., & Ortega-Vega, S. Y. (2011). El turismo alternativo como estrategia de conservación de la reserva de la biosfera de la mariposa monarca (2008-2010). Quivera Revista de Estudios Territoriales, 13, 115–133.
Rzedowsky, J. (1986). Vegetación de México. México D.F.: Limusa.
Salcido, D. M., Forister, M. L., García-Lopez, H., & Dyer, L. A. (2020). Loss of dominant caterpillar genera in a protected tropical forest. Scientific Reports, 10, 422. https://doi.org/10.1038/s41598-019-57226-9
Sánchez-Herrera, M., Forero, D., Calor, A. R., Romero, G. Q., Riyaz, M., Callisto, M. F. et al. (2024). Systematic challenges and opportunities in insect monitoring: a Global South perspective. Philosophical Transactions of the Royal Society B, 379, 20230102. https://doi.org/10.1098/rstb.2023.0102
Sánchez, M. V., Cicowiez, M., & Ortega, A. (2022). Prioritizing public investment in agriculture for post-COVID-19 recovery: a sectoral ranking for Mexico. Food Policy, 109, 102251. https://doi.org/10.1016/j.foodpol.2022.102251
Schuch, S., Bock, J., Krause, B., Wesche, K., & Schaefer, M. (2012). Long-term population trends in three grassland insect groups: a comparative analysis of 1951 and 2009. Journal of applied Entomology, 136, 321–331. https://doi.org/10.1111/j.1439-0418.2011.01645.x
Shumway, R. H., & Stoffer, D. S. (2005). Time series analysis and its applications (Springer texts in Statistics). Cham, Switzerland: Springer-Verlag.
Sims, M. J., Stanimirova, R., Raichuk, A., Neumann, M., Richter, J., Follett, F. et al. (2024). Global drivers of forest loss at 1 km resolution. Environmental Research Letters, 20, 074027. https://doi.org/10.1088/1748-9326/add606
Soroye, P., Ahmed, N., & Kerr, J. T. (2018). Opportunistic citizen science data transform understanding of species distributions, phenology, and diversity gradients for global change research. Global Change Biology, 24, 5281–5291. https://doi.org/10.1111/gcb.14358
Streitberger, M., Stuhldreher, G., Fartmann, T., Ackermann, W., Ludwig, H., Pütz, S. et al. (2024). The German insect monitoring scheme: establishment of a nationwide long-term recording of arthropods. Basic and Applied Ecology, 80, 81–91. https://doi.org/10.1016/j.baae.2024.08.004
Szabo, J. K., Vesk, P. A., Baxter, P. W., & Possingham, H. P. (2010). Regional avian species declines estimated from volunteer-collected long-term data using List Length Analysis. Ecological Applications, 20, 2157–2169. https://doi.org/10.1890/09-0877.1
Tang, B., Clark, J. S., & Gelfand, A. E. (2021). Modeling spatially biased citizen science effort through the eBird database. Environmental and Ecological Statistics, 28, 609–630. https://doi.org/10.1007/s10651-021-00508-1
Thogmartin, W. E., Diffendorfer, J. E., López-Hoffman, L., Oberhauser, K., Pleasants, J., Semmens, B. X. et al. (2017). Density estimates of monarch butterflies overwintering in central Mexico. PeerJ, 5, e3221. https://doi.org/10.7717/peerj.3221
Thomas, J. (2005). Monitoring change in the abundance and distribution of insects using butterflies and other indicator groups. Philosophical Transactions of the Royal Society B: Biological Sciences, 360, 339–357. https://doi.org/10.2307/2265653
Van Klink, R., August, T., Bas, Y., Bodesheim, P., Bonn, A., Fossøy, F. et al. (2022). Emerging technologies revolutionise insect ecology and monitoring. Trends in Ecology & Evolution, 37, 872–885. https://doi.org/10.1016/j.tree.2022.06.001
van Strien, A. J., van Swaay, C. A., van Strien-van Liempt, W. T., Poot, M. J., & Wallis De Vries, M. F. (2019). Over a century of data reveal more than 80% decline in butterflies in the Netherlands. Biological Conservation, 234, 116–122. https://doi.org/10.1016/j.biocon.2019.03.023
Vidal, O., & Rendón-Salinas, E. (2014). Dynamics and trends of overwintering colonies ofthe monarch butterfly in Mexico. Biological Conservation, 180, 165–175. http://dx.doi.org/10.1016/j.biocon.2014.09.041
Wagner, D. L., Fox, R., Salcido, D. M., & Dyer, L. A. (2021). A window to the world of global insect declines: Moth biodiversity trends are complex and heterogeneous. Proceedings of the National Academy of Sciences, 118, e2002549117. https://doi.org/10.1073/pnas.2002549117
Willott, S. (2001). Species accumulation curves and the measure of sampling effort. Journal of Applied Ecology, 38, 484–486. https://doi.org/10.1046/j.1365-2664.2001.00589.x
Zamora-Gutiérrez, V., Ortega, J., Ávila-Flores, R., Aguilar-Rodríguez, P. A., Alarcón-Montano, M., Ávila-Torresagatón, L. G. et al. (2020). The Sonozotz project: assembling an echolocation call library for bats in a megadiverse country. Ecology & Evolution, 10, 4928–4943. https://doi.org/10.1002/ece3.6245
Zylstra, E. R., Ries, L., Neupane, N., Saunders, S. P., Ramírez, M. I., Rendón-Salinas, E. et al. (2021). Changes in climate drive recent monarch butterfly dynamics. Nature Ecology & Evolution, 5, 1441–1452. https://doi.org/10.1038/s41559-021-01504-1
